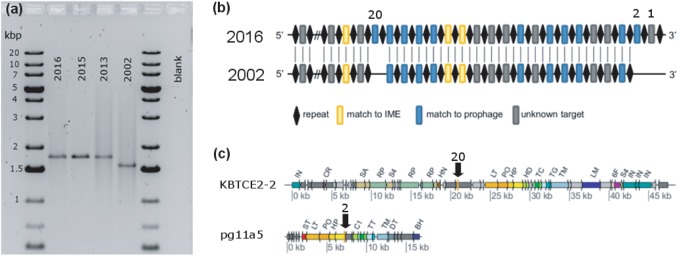
Fig. 6.
Evidence for the extension of D. mccartyi KBVC1 CRISPR array and new targets acquired. a Agarose gel showing PCR product of CRISPR array from KB-1/VC-H2 amplified from DNA extracted over time. DNA extracted in different years was stored at −80 °C. Amplified products were sequenced with internal primers; primers used are provided in Table S1, and corresponding sequences in File S1. Expected 1725 bp PCR product produced (2013–2016). Smaller 1542 bp PCR product produced from 2002 DNA. b Schematic of partial CRISPR array in 2016 compared to 2002 indicating the addition of three new spacers as determined from sequencing of PCR products. Repeats are shown as black diamonds. Blue rectangles indicate spacer match to prophage. Yellow rectangles indicate spacer match to D. mccartyi IME1. Only 24 spacers of the 41 spacers are shown in the region where new spacers were added; cross-hatches indicate where the other spacer-repeat sequences occur. New spacers are identified by spacer number (1, 2, and 20). c New spacers have best matches to two different D. mccartyi prophages indicated with black arrows. Prophages are annotated using same abbreviations as in Fig. 3 and Table S4. All spacer sequences are in supplemental File S2