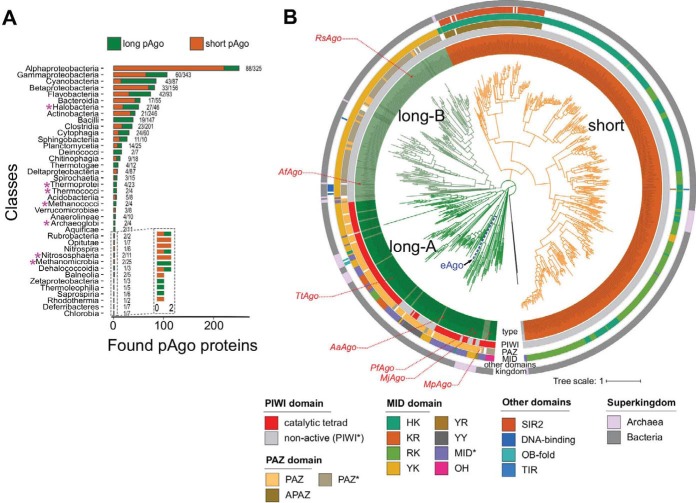
FIG 1.
Phylogenetic analysis of pAgo proteins. (A) The numbers of identified short and long pAgo proteins in the classes of Eubacteria and Archaea. Pink asterisks near the clade names indicate the archaeal classes. The numbers on the right of each bar depict the number of genera encoding pAgos versus the total number of genera with the complete or partially sequenced genomes in the corresponding class. (B) The circular phylogenetic tree of the nonredundant set of pAgos constructed based on the multiple alignment of the MID-PIWI domains. The pAgo proteins were annotated as follows, from the inner to the outer circles: the type of protein, in which long-A pAgos are colored in green (truncated variants without the PAZ domain, light green), long-B pAgos are light green (truncated variants without PAZ, green), and short pAgos are orange; the type of the PIWI domain, depending on the presence of the catalytic tetrad DEDX; the type of the 5′-end guide binding motif in the MID domain (the first two conserved residues are indicated); the presence and the type of the PAZ/APAZ domains in pAgos; and the superkingdom to which the corresponding pAgo belongs. Biochemically characterized pAgos (or the most similar redundant homologs for RsAgo and TtAgo) with resolved structures are highlighted in red. The scale bar represents the evolutionary rate calculated under the JTT+CAT evolutionary model.