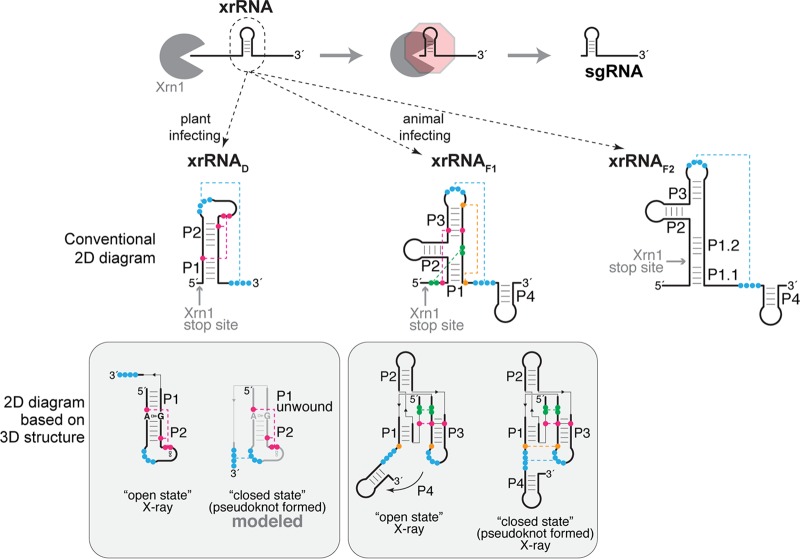
FIG 1.
An expanding repertoire of structured RNAs for blocking exoribonuclease degradation. (Top) xrRNAs adopt a three-dimensional structure that blocks the progression of 5′-to-3′ exoribonucleases such as Xrn1 (gray). In the case of flaviviruses and dianthoviruses, xrRNAs are in the 3′UTR, resulting in accumulating noncoding sgRNAs. (Middle) Secondary structure diagrams of the two classes of xrRNAs from flaviviruses (xrRNAF1 and xrRNAF2) (15, 22, 23) and of xrRNAD from dianthoviruses (26). Secondary structure features are labeled, and nucleotides involved in tertiary interactions are shown in colors connected by dashed lines (pseudoknot shown in blue). Experimentally determined Xrn1 stop sites are indicated. (Bottom) The boxes below each secondary structure contain diagrams reflecting the currently available three-dimensional structures (24–26). The A8-G33 pair is highlighted in the open state of the Sweet clover necrotic mosaic virus (SCNMV) xrRNA (far left).