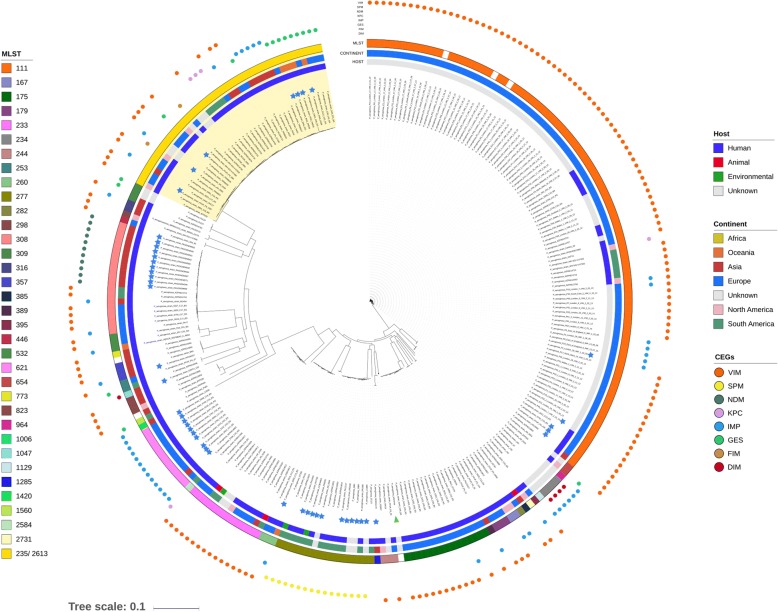
Fig. 1.
Whole-genome phylogeny of the CEG-carrying P. aeruginosa isolates. The maximum-likelihood phylogenetic tree was constructed using 146,106 single nucleotide polymorphisms (SNPs) spanning the whole genome and using the P. aeruginosa PAO1 genome (highlighted by a green triangle) as a reference. Multilocus sequence typing (MLST), continent and host data are reported on the outer-most, middle and inner-most circles, respectively. The strains belonging to a double ST profile (ST235/ST2613) are shaded yellow. Blue stars point out P. aeruginosa strains for which a CEG-harbouring ICE was predicted. The P. aeruginosa AR_0356 genome (accession number CP027169.1) was removed from the tree since it corresponds to a strain of which host and origin are unknown. The phylogenetic distance from the tree root to this genome is 1 (calculated with the tree scale). The Newick format file for the original tree is included in the Additional file 3