Abstract
The Dmrt (doublesex and mab-3-related transcription factor) genes are transcription factors crucial for sex determination and sexual differentiation. In some social insects, doublesex (dsx) exhibits widespread caste-specific expression across different tissues and developmental stages and has been suggested as a candidate gene for regulating division of labor in social insects. We therefore conducted a molecular evolution analysis of the Dmrt gene family in 20 ants. We found that the insect-specific oligomerization domain of DSX, oligomerization domain 2, was absent in all ants, except for the two phylogenetically basal ant species (Ponerinae), whose social structure and organization resemble the presumed ancestral condition in ants. Phylogenetic reconstruction and selection analysis revealed that dsx evolved faster than the other three members of the Dmrt family. We found evidence for positive selection for dsx in the ant subfamilies with more advanced social organization (Myrmicinae and Formicinae), but not in the Ponerinae. Furthermore, we detected expression of two Dmrt genes, dsx and DMRT11E, in adult ants, and found a clear male-biased expression pattern of dsx in most species for which data are available. Interestingly, we did not detect male-biased expression of dsx in the two ant species that possess a genetic caste determination system. These results possibly suggest an association between the evolution of dsx and social organization as well as reproductive division of labor in ants.
Keywords: sex determination, caste determination, eusocial insect, transcription factor, transcriptome, alternative splicing
Introduction
Ants (Hymenoptera, Formicidae), which are one of the most successful eusocial taxa (Gadau et al. 2012), display large variation in the complexity of social organization, ranging from some “primitive” ponerine species, whose workers have full reproductive potential, to species where workers are totally sterile and exhibit morphologically distinct subcastes (Hölldobler and Wilson 1990, 2009). In the majority of ants, only queens can mate and reproduce, whereas workers are incapable of mating, because they lack functional spermathecae (Bourke 1988).
The genetic pathways regulating the determination of the queen and worker castes include the insulin/insulin-like growth signaling (IIS), target of rapamycin, epidermal growth factor receptor, juvenile hormone (JH), and vitellogenin. External environmental stimuli, such as nutrition and temperature, are important inputs into these integrative pathways. However, the downstream effector pathways connecting these regulators to growth and differentiation remain largely unknown (reviewed in Corona et al. [2016]).
In solitary insects, the sex-differentiation gene doublesex (dsx) links development to nutrition and sex. For example, dsx regulates sex-specific mandible growth via JH signaling in stag beetles (Gotoh et al. 2014). Drosophila melanogaster dsx was the first Dmrt gene (doublesex and mab-3-related transcription factor) identified. dsx encodes two sex-specific splice variants that regulate transcription of target genes to trigger sexual differentiation (Nagoshi et al. 1988). Both the male- and female-specific protein products, DSXM and DSXF, share an N-terminal DM domain, which consists of a zinc finger-related DNA-binding domain and an oligomerization domain (OD1) (Erdman and Burtis 1993; An et al. 1996). Dmrt family members are defined by the presence of the DM domain and have been found in almost all eumetazoans (Wexler et al. 2014). There are eight Dmrt genes in Homo sapiens and 11 in Caenorhabditis elegans (Ottolenghi et al. 2002; Volff et al. 2003). In insects, four Dmrt genes are known (DMRT11E, DMRT93B, DMRT99B, and dsx) (Volff et al. 2003).
The C-termini of DSXM and DSXF contain another oligomerization domain, OD2, which consists of both a shared oligomerization sequence and sex-specific sequences. DSX forms dimers to bind DNA and regulate the transcription of target genes. The dimerization of the monomer DSXs is caused by OD1 and OD2 independently (An et al. 1996; ErdmAn et al. 1996). DSXF and DSXM bind to the same dimorphic cis-regulatory-element, but the sex-specific OD2 domains may recruit different sets of regulatory proteins, thereby regulating sex-specific expression of target genes (reviewed in Verhulst and van de Zande [2015]). Consequently, OD2 is important to the double-switch function of DSX in regulating sexual differentiation.
As a pleiotropic transcription factor, dsx controls a wide range of downstream patterning genes responsible for the development of sex-related traits. Among them, the most ancestral function of dsx was likely the determination of gonadal sex (Matson and Zarkower 2012), including regulating the differentiation of genital structures, such as the spermathecae (Sanchez et al. 2001). dsx also regulates biosynthesis of pheromone (Shirangi et al. 2009) and sexual behaviors (Rideout et al. 2010). Given the important roles dsx plays in multiple aspects of reproduction, dsx may be involved in the polyphenism of eusocial insects between and within sexes. In line with this view, several studies have revealed caste-specific expression of dsx across different tissues in the honeybee Apis mellifera or development stages in the ant Cardiocondyla obscurior and hypothesized that dsx may regulate polyphenism in social insects (Johnson and Jasper 2016; Klein et al. 2016).
Compared with other eusocial insects, the gradient of social complexity (Hölldobler and Wilson 1990, 2009) along with their rich taxonomic diversity makes ants an ideal model system for investigating the evolution of caste polyphenism. In this study, we annotated Dmrt genes in 20 ant species and predicted their conserved domains. Overall, the OD2 domain is present in some phylogenetically basal ants that show weak reproductive division of labor but is lost in several more advance ant lineages. We also found that the evolutionary rate of dsx is relatively high among the four Dmrt genes, especially in ants showing more advanced social organizations. These results suggest that the coding sequences of dsx genes are diverging congruently with variation in social organization. We then used transcriptome data to analyze sex- or caste-biased gene expression and alternative splicing of Dmrt genes, and found that transcription of dsx was higher in males for most examined Hymenopteran species, except for two Myrmicinae ants, which are known for their peculiar caste-determination mechanisms (Fournier et al. 2005; Ohkawara et al. 2006). Furthermore, the exon-usage of dsx in these two Myrmicinae species shows significant differences not only between sexes but also among castes. Taken together, the evolutionary and expression analyses provide multiple lines of evidence that the evolutionary characteristics of dsx map well to the evolution of the caste determination in ants.
Materials and Methods
Identification of Dmrt Genes and Phylogenetic Reconstruction
The annotated genes and genome assemblies of 48 insect species, including 39 Hymenopteran species (20 ants and 19 nonant Hymenoptera) who were the only Hymenopteran species with available genomes at the start of this study, and nine representatives for non-Hymenopteran species whose genomes were also available. Besides, six Crustaceans whose Dmrt genes had been reported (Kato et al. 2008, 2011; Toyota et al. 2013; Zhang et al. 2014) were also included in our research as outgroups to the insects. Our analysis was focused on 20 ant species. Four D. melanogaster Dmrt genes (Dmel_DMRT11E, DMRT93B, DMRT99B, and dsx) were used as queries in the BLASTp search (Altschul 1997), with an E-value cutoff of 10−5. We reiterated a tBLASTn search in the genome assemblies using each candidate Dmrt sequences as queries, with an E-value cutoff of 10−4. Then, a hidden Markov model (HMM)-based tool FGENE SH+ (Buschinger 1968) in the SOFTBERRY package (http://linux1.softberry.com; Last Accessed December 7, 2018) was used to improve the exon–intron boundaries. Gene fragments without a full DM domain or with an open reading frame (ORF) length <500 bp were categorized as pseudogenes.
Eighty-two amino acid sequences of Dmrt genes from 20 ant species and two outgroup Vespidae species were aligned using MAFFT version 7 (Katoh and Standley 2013). The resulting alignment was analyzed using PROTTEST3 to predict the bestfit substitution model of molecular evolution (Darriba et al. 2011). The phylogenetic tree was constructed using the PHYML3.0 program (Criscuolo 2011) with the Jones–Taylor–Thornton model, gamma distribution of rates, optimized equilibrium frequencies and invariant site categories, with 100 bootstrap replications.
Selection Analysis
PAML analyses were performed based on the species tree shown in figure 1. The ratios of nonsynonymous (dN) and synonymous (dS) substitution rates (ω = dN/dS) were evaluated using CODEML in PAML version 4.5 (Yang 2007). For each Dmrt subfamily, tests using different site models were conducted: the basic model (M0: one ratio) assuming ω is invariable among sites and branches (Goldman and Yang 1994), the site-specific models assuming ω varies among sites but not branches, including M1a (neutral), M2a (selection), M3 (discrete), M7 (beta), M8 (beta & ω > 1), and M8a (beta & ω = 1) (Yang et al. 2000). Pairwise comparisons of M0 versus M3, M1a versus M2a, M7 versus M8, and M8a versus M8 were used to perform likelihood ratio tests (LRTs). For dsx genes, subsequently analysis using the branch-site model, allowing ω to vary both among sites and branches (model = 2, NSsites = 2) (Zhang et al. 2005), was tested (df = 1) against a null model (fixed ω of 1). LRTs using the pairwise comparisons of branch-site model versus branch-site null model were performed. Significance tests were performed using the χ2 distribution. After, if the LRT was significant, a Bayes empirical Bayes (BEB) analysis was conducted to identify positively selected sites (P > 0.95).
Fig. 1.
—Domain organization of ant DMRTs. Domains of DMRT proteins for 20 ant species, two outgroup Vespidae species, and D. melanogaster. Full-length proteins for each of the four DMRT genes are shown as hollow boxes in four distinct colors. DM, DMA, and OD2 domains are illustrated as solid boxes in black, gray, and red, respectively. OD2-like domains found in H. saltator and Di. quadriceps are shown as hashed red boxes. The branches belonging to the subfamilies of Myrmicinae, Formicinae, and Ponerinae are colored in blue, green, and yellow, respectively. The phylogenetic relationship is based on Moreau and Bell (2013) and Ward et al. (2015). The plus signs indicate the six branches undergone positive selection.
Gene Expression Analysis
Solenopsis invicta colonies were collected in Guangdong Province, South China, on October 16, 2017. We selected a colony whose monogyne status was verified by using a polymerase chain reaction protocol previously described (Krieger and Ross 2002). After being reared in the lab for 2 weeks, three biological replicated samples were chosen from the same three colonies. Total RNA was isolated from whole-body adults of three castes, (workers, egg-laying queens, and presumptive haploid males) separately, with three biological replicates for each caste. For the RNA extraction, the TransZol Up Plus RNA Kit (Transgen Biotech, Beijing, China) was used in accordance with the manufacturer’s instructions. These mRNA samples were then submitted to the Berry Genomics Company (Beijing, China) for mRNA-seq library construction and sequencing. The sequencing was conducted using an Illumina HiSeq 4000 (Illumina, San Diego, CA) platform, with a paired-end module (PE150 mode). The transcriptome data of S. invicta have been deposited to the NCBI's Sequence Read Archive (SRA) database (http://www.ncbi.nlm.nih.gov/Traces/sra; Last Accessed December 7, 2018) and given the accession number SRP143788. Fastq files for other species (supplementary table S1, Supplementary Material online) were downloaded from the SRA database. All the RNA-sequencing (RNA-seq) data were then mapped against their assembled genome sequences using Tophat (Trapnell et al. 2009) with default parameters. To estimate the transcript level abundance in each sample, we calculated fragments per kilobase of transcript per million fragments mapped (FPKM) using CUFFLINKS (Trapnell et al. 2012). Genes with FPKM >10 were used for subsequent analyses. For the dsx gene which expressed multiple alternatively spliced transcripts, FPKMs for each of the transcripts were calculated separately by CUFFLINKS and were then summed up to evaluate the expression level of the gene. Only gene isoforms with a DM domain were analyzed. In order to control for the differences caused by the different sequencing protocols, the expression levels of the housekeeping genes ribosomal protein L18, elongation factor 1, and NADH dehydrogenase were also determined for each caste, using the same method.
Evidence of differential exon-usage in the dsx and DMRT11E genes was tested using the DEXSeq package’s (version 1.22.0) “exon-centric” differential splicing analysis (Li et al. 2015) in R (R Core Team 2000). Prior to analysis, transcript models produced by CUFFLINKS were used as input for the script to prepare the annotation used in DEXSeq. An LRT was performed, comparing the full model (∼sample + exon + caste:exon) and reduced model (∼sample + exon), to detect differences in exon-usage.
Results
Gene Annotation
We searched the genome assemblies and the predicted gene sets from 20 ant species and 19 other Hymenopteran species (supplementary table S2, Supplementary Material online) for Dmrt homologs. We combined these Hymenopteran Dmrt genes with 45 additional Dmrts from nine representatives of other insect orders and six Crustaceans as insect outgroup and then built a phylogeny. Examination of the Dmrt tree topology suggested a diversification of the four Dmrt subfamilies prior to the diversification of Crustacea and Hexapoda (supplementary fig. S1, Supplementary Material online). We used a nomenclature for insect Dmrt genes according to the Dmrt genes in D. melanogaster (DMRT11E, DMRT93B, DMRT99B, and dsx) (Volff et al. 2003).
In Hymenoptera, lineage-specific gene duplication and loss are rare (supplementary table S2, Supplementary Material online). We annotated partial gene fragments on separated scaffolds as pseudogenes, but these are likely genome assembly artifacts. Aside from these artificial pseudogenes, our analysis suggests that all the clear cases of expansion and contraction of Dmrt genes took place in the lineages giving rise to Atta cephalotes, Pseudomyrmex gracilis, and Wasmannia auropunctata (bold in supplementary table S2, Supplementary Material online).
Loss of OD2 Domain in Ants
We used the NCBI’s Conserved Domain Detector (CDD) (Marchler-Bauer et al. 2015) to identify conserved domains of the Dmrt genes. All the examined genes had a DM domain that is characteristic for the Dmrt gene family. All predicted orthologs of DMRT11E, DMRT93B, and DMRT99B shared the same domain organization as D. melanogaster, including the presence of a DMA domain in DMRT93B and DMRT99B (Ottolenghi et al. 2002; Volff et al. 2003).
Interestingly, the conserved domain composition of DSX proteins was more variable (fig. 1 and supplementary fig. S1, Supplementary Material online). We did not detect the OD2 domain in any ant DSX, even when we relaxed the E-value threshold to 0.01. To search for remote homologs, we further queried the 20 ant genomic data sets via BLASTn/tBLASTn (E-value = 1.0) with the OD2 sequence of A. mellifera. There was evidence for the presence of a DSX OD2 domain only in Harpegnathos saltator and Dinoponera quadriceps, both of which are ponerine ants. The alignment of Hsal_DSX and Dqua_DSX with other Hymenopteran DSXs indicated an exclusive 5-amino acid insertion in the middle of the OD2 domain in these two ponerine ant sequences (supplementary fig. S2, Supplementary Material online). When aligned pairwise with other Hymenopteran DSXs, the average sequence identity was lower for Hsal_DSX and Dqua_DSX (0.23) than other Hymenopteran DSXs (0.42). The high level of sequence divergence was likely the reason why we could not detect the OD2 domain using the CDD database.
To ascertain that the loss of OD2 domain was not an assembly artifact, we queried the short-read sequencing data of the 20 ants with the seed alignments for the OD2 (or OD2-like) domain sequences of D. melanogaster, H.saltator, and Di.quadriceps via SRA-BLAST (Camacho et al. 2009). In addition, an HMM profile (HMMER version 3.1) (Eddy 1998) was created with the same alignment of OD2 domain sequences as used in the SRA-BLAST search, and a profile nHMMER search was carried out against the genome databases of the 20 ants, with a threshold E-value of 0.01. Both of these searches failed to detect OD2 domain homologs in the other 18 ant species.
We further examined the Hymenopteran dsx genes with and without the OD2 domain for any additional structural differences. We found that the 5′-terminal sequences of dsx (DM domain coding sequences) are conserved among all the species. In contrast, the 3′-terminal sequences of dsx are quite variable, both for ants (without OD2) and for nonant species (with OD2). We have aligned the OD2 domain of DSX in different Hymenopteran species (with OD2 for nonant species, and OD2-like domain for the two ponerine ants), as shown in figure S2, Supplementary Material online. But it seems impossible to align the 3′-terminal sequences of dsx for “higher” ants (without OD2), because the sequence similarity is too low.
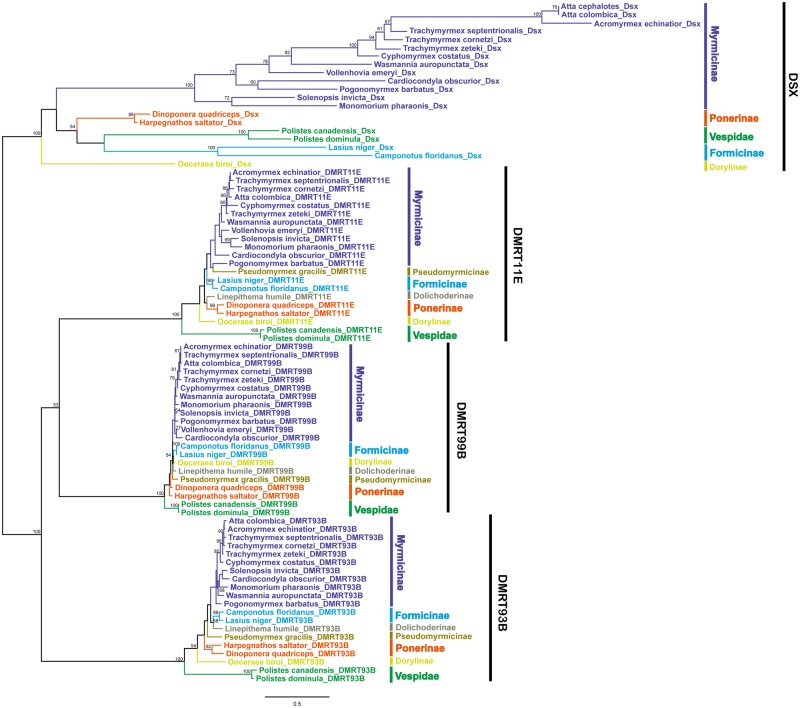
Gene Phylogeny
Our phylogenetic analysis (fig. 2) revealed that the Vespidae DMRT11E, DMRT93B, and DMRT99B genes form outgroups to the Formicidae homologs, which is consistent with the taxonomic relationship of these species. However, the pattern was strikingly different for dsx, with the two Vespidae dsx not forming an ancestral lineage to the Formicidae lineages. After conducting a phylogenetic analysis with only DSX sequences, the two Vespidae dsx sequences rooted basal to the Formicidae dsx sequences (supplementary fig. S3, Supplementary Material online), suggesting that the ingroup placement of Vesipdae dsx in figure 2 was caused by long-branch attraction (Felsenstein 1978). The branch lengths separating taxa in the dsx clade were long, indicating high evolutionary divergence of dsx across the different species included in the analysis. A phylogenetic tree built with only the DM domain showed a similar topological structure (supplementary fig. S4, Supplementary Material online), indicating that the evolutionary rate of the DM domain, which is thought to be conserved among Dmrt homologs, is extremely high in dsx. Taken together, these results indicate that the dsx genes have the fastest rate of evolution among the four Dmrt genes.
Fig. 2.
—Maximum-likelihood phylogenetic analysis of the deduced amino acid sequences of Dmrt homologs in ants and two vespids using PHYML. Bootstrap values are indicated for each node (of 100 resamplings).
dsx Genes of Ants Have Evolved under Positive Selection
The finding that the tree topology of the Dmrt genes (fig. 2) does not mirror that of the species tree (fig. 1) hinted that the Dmrt genes might undergo lineage-specific selection in ants. We further estimated the ratios of nonsynonymous (dN) and synonymous (dS) substitution rates for the four Dmrt genes using the PAML package (Yang 2007). Because of the extreme divergence of the C-terminus of DSX, it was difficult to determine homologous codon positions outside the DM domain. So only the coding sequence for the N-terminus of the DSX protein, which includes the DM domain, was used in the PAML analysis. We analyzed the four Dmrt genes separately under multiple site-specific models, which allow ω = dN/dS to vary among sites (table 1 and supplementary tables S3–S5, Supplementary Material online). Under the basic model (M0), which assumes ω to be invariable among sites and branches (Goldman and Yang 1994), dsx showed a higher ω (0.62) than the other three Dmrt genes (0.05–0.16). To test whether ω values among sites were variable, we compared M0 with the discrete model (M3) using LRTs. Another two LRTs, which compared M1a (neutral) with M2a (selection), and M7 (beta) with M8 (beta & ω), were also conducted. For all four Dmrt genes, M3 was a significantly better fit than M0, indicating that ω is different among sites. Using a BEB approach to identify positively selected sites (in the case of models M2a and M8), we found no positively selected sites for DMRT11E, DMRT93B, and DMRT99B (supplementary tables S3–S5, Supplementary Material online). By contrast, multiple amino acid sites of dsx (12 sites in M2a and 10 sites in M8) apparently underwent adaptive evolution, and LRTs performed were significant for both of these models (table 1).
Table 1.
Results of Site-Specific Positive Selection Analysis of the dsx Gene
| Models | P | ln L | Parameters | PS Sites | Significant after LRT Correction |
|---|---|---|---|---|---|
| M0: One ratio | 40 | −4,240.610423 | ω = 0.62265 | None | — |
| M1a: neutral | 41 | −4,131.232251 | p0 = 0.33583, p1 = 0.66417 | Not allowed | — |
| ω0 = 0.16958 , ω1 = 1.00000 | |||||
| M2a: selection | 43 | −4,107.690927 | p0 = 0.19604, p1 = 0.53092 | BEB | M1a–M2a: yes |
| (p2 = 0.27304), ω0 = 0.10031 | 6 > 0.95 | ||||
| ω1 = 1.00000, ω2 = 3.19922 | 6 > 0.99 | ||||
| M3: discrete | 44 | −4,097.544406 | p0 = 0.04743 (p1 = 0.52106) | NEB | M0–M3: yes |
| p2 = 0.43152, ω0 = 0.00510 | 5 > 0.95 | ||||
| ω1 = 0.51352, ω2 = 1.46193 | 20 > 0.99 | ||||
| M7: beta | 41 | −4,110.962547 | p = 0.46067, q = 0.25504 | Not allowed | — |
| M8: beta & ω >1 | 43 | −4,094.79483 | p0 = 0.74246, p = 0.49554 | BEB | M7–M8: yes |
| q = 0.36279 (p1 = 0.25754), | 5 > 0.95 | ||||
| ω = 2.53805 | 5 > 0.99 | ||||
| M8a: beta & ω = 1 | 42 | −4,110.28144 | p0 = 0.57543, p = 0.63016 | Not allowed | M8a–M8: yes |
| q = 1.07704 (p1 = 0.42457), ω = 1.00000 |
Note.—The table gives the number of sites predicted to be under positive selection for each site-specific model. Abbreviations are as follows. Models, site-specific models; P, number of parameters; ln L, log likelihood scores; p, proportion of sites under a particular omega value; ω, ratio of nonsynonymous to synonymous substitution rates; PS sites, number of positively selected sites; NEB, naive empirical Bayes analysis.
The alignment of the DSX N-terminal coding sequences was further analyzed using the branch-site model A in PAML, which allows ω to vary both among branches and sites (table 2). dsx genes from each of the ant species were tested as foreground branches. Positively selected sites could be detected in six species, four of which belong to the Myrmicinae subfamily and the other two to the Formicinae subfamily (fig. 1 and table 2). LRT analysis indicated that the unconstrained model (model HA) fitted the data sets significantly better than the model fixing ω for the foreground branches at 1 (model H0) in all six cases. Interestingly, no positively selected sites could be found in Ponerinae ants (table 2). This suggests that among the four Dmrt genes, positive selection occurred only in dsx, and that this occurred only in the two “higher” ant subfamilies, but not in the “primitive” Ponerinae.
Table 2.
Tests for Positive Selection on dsx Homologs in Ants Using the PAML Branch-Site Model A
| Branch | H0: p2a (ω2 = 1) | HA: p2a, ω2 | PS Sites (BEB) | Significant after LRT Correction |
|---|---|---|---|---|
| Myrmicinae | ||||
| Atta colombica | 0.00 | 0.02, 1.00 | None | No |
| Atta cephalotes | 0.01 | 0.06, 2.61 | None | No |
| Acromyrmex echinatior | 0.01 | 0.10, 999.00 | 4 > 0.95 | Yes |
| Trachymyrmex septentrionalis | 0.07 | 0.07, 1.86 | None | No |
| Trachymyrmex cornetzi | 0.05 | 0.04, 5.37 | None | No |
| Trachymyrmex zeteki | 0.10 | 0.24, 1.00 | 1 > 0.95 1 > 0.99 |
Yes |
| Cyphomyrmex costatus | 0.00 | 0.03, 17.94 | None | Yes |
| Wasmannia auropunctata | 0.08 | 0.08, 22.14 | 2 > 0.95 1 > 0.99 |
Yes |
| Vollenhovia emeryi | 0.00 | 0.01, 18.18 | None | No |
| Cardiocondyla obscurior | 0.00 | 0.03, 207.50 | None | No |
| Solenopsis invicta | 0.06 | 0.07, 88.31 | 1 > 0.95 1 > 0.99 |
Yes |
| Monomorium pharaonic | 0.01 | 0.03, 13.05 | None | No |
| Pogonomyrmex barbatus | 0.00 | 0.00, 1.00 | None | No |
| Formicinae | ||||
| Lasius niger | 0.13 | 0.09, 999.00 | 1 > 0.95 | Yes |
| Camponotus floridanus | 0.11 | 0.13, 51.08 | 1 > 0.95 2 > 0.99 |
Yes |
| Cerapachyinae | ||||
| Ooceraea biroi | 0.02 | 0.04, 133.61 | None | No |
| Ponerinae | ||||
| Dinoponera quadriceps | 0.00 | 0.00, 1.00 | None | No |
| Harpegnathos saltator | 0.00 | 0.00, 1.00 | None | No |
Overall Male-Biased Expression of dsx in Ants
To test whether Dmrt genes show sex- or caste-specific gene expression in ants, we gathered transcriptomic data for five ant species (Camponotus floridanus, H. saltator, S. invicta, Vollenhovia emeryi, and W. auropunctata). The expression levels of the housekeeping genes as ribosomal protein L18, elongation factor 1, and NADH dehydrogenase, which have proven to be valid controls in Hymenopteran species (Scharlaken et al. 2008; Cheng et al. 2013; Helmkampf et al. 2015; Livramento et al. 2018), were used to normalize gene expression across different samples. The expression of these reference genes was consistent within species (coefficients of variation ≦ 0.74), making them suitable reference genes to compare Dmrt gene expression levels within species (supplementary fig. S5, Supplementary Material online).
There were common features and also interesting species-specific differences in Dmrt expression profiles. dsx and DMRT11E showed the strongest expression in ants, with DMRT11E expression levels being generally lower than dsx. The expression levels of the other two Dmrt genes were too low to be detected in most cases (fig. 3). In contrast, dsx and DMRT99B, but not DMRT11E, were expressed at higher levels than the other two Dmrt genes in three other nonant Hymenopterans (supplementary fig. S6, Supplementary Material online). In summary, among the four Dmrt genes, dsx was widely expressed in all Hymenopteran species included in this study, whereas DMRT11E was expressed only in the ant lineage.
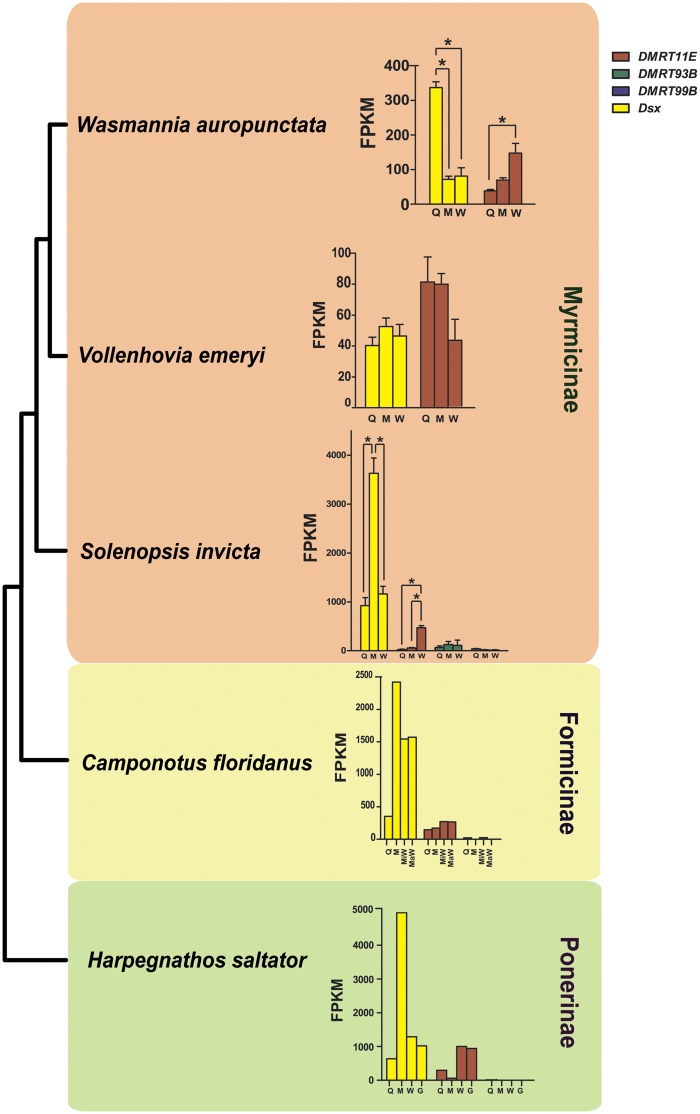
Fig. 3.
—Sex- and caste-specific gene expression levels of Dmrt genes in five ant species. Gene expression levels are shown in FPKM. The phylogenetic relationship is based on Moreau and Bell (2013). For species with at least three biological replicates (W. auropunctata, V. emeryi, and S. invicta), error bars represent standard error of the mean (SEM), and asterisks indicate P-value from a one-way ANOVA (P < 0.05), followed by the Tukey’s HSD post hoc test. Q, queen; M, male; W, worker; MiW, minor worker; MaW, major worker (Cam. floridanus has two nonreproductive castes); G, gamergate (workers having the ability to replace the queen when the queen dies [Peeters et al. 2000]). Genes that are not expressed in any caste are not shown in the graph.
We then tested whether Dmrt expression was sex- or caste-biased in ants. We detected male-biased expression of dsx in three out of the five ant species (fig. 3). The expression of dsx was significantly higher in males than in queens and workers in the fire ant S. invicta (one-way ANOVA, P-value < 0.01, followed by the Tukey’s Honestly Significant Difference [HSD] post hoc test). dsx gene expression was similarly male-biased in Cam. floridanus and H. saltator as well as the three nonant Hymenopteran species examined, although statistical tests were not possible in these species (fig. 3 and supplementary fig. S6, Supplementary Material online).
The other two ant species did not display male-biased expression of dsx. In V. emeryi, dsx and DMRT11E were expressed in all three castes, resembling the expression profiles of the other four ants (fig. 3). However, the expression levels of dsx and DMRT11E did not differ significantly among the different castes (one-way ANOVA, dsx: P-value = 0.40; DMRT11E: P-value = 0.10). No difference in dsx expression levels was detected even when queens and workers were pooled as females in comparison to males (t-test, P-value = 0.24). In addition, DMRT11E expression levels were significantly higher than dsx in queens (t-test, P-value = 0.04) and males (t-test, P-value = 0.01). In W. auropunctata, dsx expression was significantly higher in queens than both males and workers (one-way ANOVA, P-value < 0.01, followed by the Tukey’s HSD post hoc test), and DMRT11E expression was significantly higher in workers compared with queens (one-way ANOVA, P-value = 0.02, followed by the Tukey’s HSD post hoc test). The queen-biased rather than male-biased expression of dsx stands in contrast to the other species (fig. 3) and is distinctive among all other insects included in the analysis.
dsx Has Different Exon-Usage Patterns Compared with DMRT11E in Ants
The 3′-sequence of dsx, which includes the OD2 domain, is alternatively spliced in a sex-specific manner in other insects (Baker and Wolfner 1988; Cho et al. 2007). We were curious as to whether ant dsx genes were alternatively spliced in the condition of losing OD2. We applied DEXSeq to the RNA-seq data to test for differential usage of dsx and DMRT11E exons caused by the differentiation of castes. Common dsx transcripts that are both expressed in females and males have been found in several other insects (Cho et al. 2007; Duan et al. 2013), and there are also some nonsense splice forms that are possibly degraded by nonstop-mediated decay (Duan et al. 2013). So with the existence of the common transcripts and the nonsense transcripts, the expression patterns of dsx exons detected by DEXseq were expected to overlap between the two sexes or among castes. But still, the specific spliced exons could be deduced from the significant difference of the exon usage abundance among the three castes. Unexpectedly, we found significant differential usage of dsx exons in different castes for all three ant species investigated (S. invicta, V. emeryi, and W. auropunctata; fig. 4 and supplementary table S6, Supplementary Material online). In contrast, there was no evidence of caste-specific alternative splicing for the DMRT11E ORFs (supplementary fig. S7, Supplementary Material online).
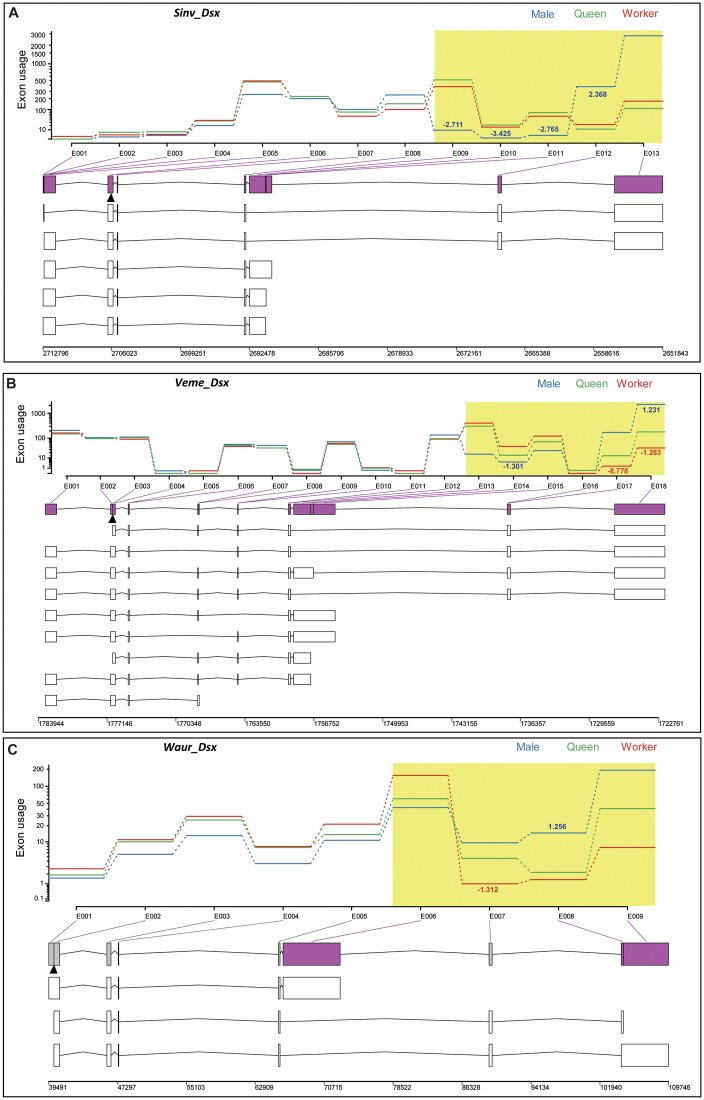
Fig. 4.
—Estimated exon usage of dsx in S. invicta (A), V. emeryi (B), and W. auropunctata (C). Exon usage is color coded for the three castes. Annotated transcript models are shown below for each species, with boxes representing exons and lines for introns. Significant differences in exon usage are shown in purple (P-value < 0.05; supplementary table S6, Supplementary Material online). The positions of start codons are indicated by black arrowheads. The predicted alternatively spliced C-terminus starting from the sex-specific skipped exon is highlighted in yellow. In these regions, exon-usage changes (absolute value of log2 fold-changes, >1) of male-to-queen and worker-to-queen are shown as blue and red numbers, respectively.
In the transcript models deduced by DEXSeq, we observed that caste-specific alternative splicing events that affect ORFs occur predominantly in the 3′-ends of dsx via caste-specific exon skipping, resulting in the production of alternative transcripts containing different distal exons (fig. 4). The patterns of alternative splicing were similar for the three species, with the skipped exons (exons 9–11 in Sinv_dsx; exons 13–15 in Veme_dsx; exon 6 in Waur_dsx) showing less expression in males than in either the queens or workers, and the distal exons (Sinv_dsx, exons 12–13; Veme_dsx, exons 16–18; Waur_dsx, exons 7–9) showing the highest expression in males among the three castes (statistical analyses are given in supplementary table S6, Supplementary Material online). These results are consistent with the conserved splicing pattern for insect dsx: Female-specific exons are skipped in males, and the male-specific transcripts are more extended at the 3′-terminus. For Sinv_dsx, there were no significant expression differences for any exons between queens and workers. In contrast, in Veme_dsx and Waur_dsx, exon usage between queens and workers for some exons was different (Veme_dsx, exons 17 and 18; Waur_dsx, exon 7; fig. 4 and supplementary table S6, Supplementary Material online).
Taken together, our results suggest that although dsx lacks the OD2 domain, it is still alternative spliced in ants, producing different C-terminal sequences with no similarity to known proteins. Furthermore, it is notable that both Veme_dsx and Waur_dsx exhibit caste-specific splicing between the two female castes.
Discussion
Evolution of Dmrt Genes in Ants
The Dmrt gene family is conserved among metazoans (Bellefroid et al. 2013). Our survey of four Dmrt genes in Hymenopterans by annotation and phylogenetic analyses revealed variation in the degree of conservation for Dmrt genes. All four Dmrt genes were primarily comprised single-copy genes, but there may be more gene gains and losses in ants than other Hymenopteran species.
Phylogenetic analyses of dsx in ants and vespids revealed long branch lengths compared with the other three Dmrt genes, indicating that dsx experienced the highest evolutionary rate among the four Dmrt genes. Accordingly, a selective pressure test revealed positively selected sites in dsx but not in the three other Dmrt genes. Moreover, the evolutionary rate of dsx genes varies in different ant subfamilies. PAML results suggest no positive selection on dsx in the phylogenetically basal subfamily Ponerinae but positive selection in the “higher ant” subfamilies Myrmicinae and Formicinae. Given that most Myrmicinae and Formicinae display a clear reproductive division of labor and a sophisticated communication systems (Hölldobler and Wilson 1990, 2009), in contrast to the Ponerinae, this raised the possibility of a role of dsx in the evolution of complex social organization.
Ant dsx Genes Are Alternatively Spliced despite Losing OD2
In our research, sequences homologous to the OD2 domain were only found in the Poneroid clade, a basal lineage of ants (Moreau and Bell 2013). Given that the OD2 domain was also present in the outgroup Vespidae, we propose that the OD2 domain was lost prior to the early radiations of the two advanced ant subfamilies (Myrmicinae and Formicinae) (Moreau and Bell 2013). Lineage-specific losses of OD2 have also been reported in Hemiptera, Embioptera, Isoptera, and Blattaria, which are hemitabolous insects (Geuverink and Beukeboom 2014; Price et al. 2015). However, there was no previous evidence of the OD2 domain being lost in holometabola.
The OD1 and OD2 domains in DSX promote oligomerization independently. OD1 and DNA-binding domain, which together compose the DM domain, are often present in all splice variants of DSX and together form a dimeric DNA-binding unit (An et al. 1996; ErdmAn et al. 1996). OD1 dimerization is important for DSX to bind to DNA, and OD2 provides additional dimerization energy, promoting and stabilizing dimeric interactions between DSX monomers (Cho and Wensink 1998).
The lack of OD2 in these ants may indicate that DSX in these species interacts with target DNAs in a monomeric form. However, although it has been reported that DSX could bind to DNA in vitro as monomer (Zhu et al. 2000), DSX binds to target DNA as dimer predominantly under conditions containing enough free DNA (Cho and Wensink 1996; Zhu et al. 2000). An alternative hypothesis is that the ant DSXs may bind to DNA in a dimeric form using the weak oligomerization energy provided by OD1. But the OD1–OD1 interaction alone may be insufficient to maintain an optimal dimeric structure, reducing DNA-binding affinity and specificity. Further studies are needed to investigate the consequence of the absence of the OD2 domain in DSX on its functional structure.
OD2 has other important roles than dimerization. The sex-specific OD2 confers upon Drosophila DSX the ability to regulate sex-specific gene expression. OD2 dimerization mediates the recruitment of other transcriptional cofactors such as Intersex (Garrett-Engele et al. 2002), increases DNA binding of the DM domain (Cho and Wensink 1998), and enhances binding site recognition (Cho and Wensink 1997). Changes that abolish OD2 dimerization result in an intersexual phenotype (ErdmAn et al. 1996). As a result, it seems possible that the absence of OD2 in ants may alter the specific transcriptional regulatory activity of DSX on its target genes.
However, although the dsx of higher ants lacks OD2, it is still alternatively spliced. There are sex- and caste-specific exon usages of ant dsx genes, without the OD2 domain. These results suggest that novel sex- or caste-specific DSX C-termini are generated by alternative splicing in these ants. Rather than completely losing their function, ant DSXs could have retained the ability to recruit different regulatory complexes in different sexes or castes. More functional experiments are needed to explore how DSX protein–protein interactions may be different in ants.
Unusual Expression of dsx in Two Genetic Caste Determination Ants
Our comparative transcriptomic analysis across eight Hymenopteran species revealed a male-biased expression pattern of dsx in six species. Male-biased expression of dsx was also found in the ant C. obscurior (Klein et al. 2016). Homologs of dsx in arthropods outside of Hexapoda, such as water fleas and mites, also display male-biased expression (Kato et al. 2011; Pomerantz and Hoy 2015). However, the ants W. auropunctata and V. emeryi did not show the typical male-biased expression. In contrast to S. invicta and other well-studied insects that only generate different dsx isoforms between sexes, these two species express dsx transcripts differently both between sexes (queen and male) and among castes (worker and queen).
The distinctive dsx expression patterns in W. auropunctata and V. emervi may be related to their special caste determination system. While most ants have an environmental caste determination system where the queen and worker castes are determined by environmental stimuli, W. auropunctata and V. emeryi have a genetic caste determination system, where caste differentiation is predicted by the individual’s genotype (Schwander et al. 2010). In these two species, queens are produced by ameiotic parthenogenesis while workers are produced through normal sexual reproduction. Moreover, in both species males are produced clonally from their father, possibly as a result of the elimination of the maternal genome once the eggs have been fertilized (Fournier et al. 2005; Ohkawara et al. 2006). It would be of interest to study whether the noncanonical expression patterns of dsx in these two species are associated with their unusual mode of caste determination.
dsx May Be Related to Caste Differentiation in Ants
Another line of evidence that dsx may be associated with caste differentiation in ants is the correlation between dsx evolutionary patterns and the level of social organization. In contrast to most other ants, the workers of the phylogenetically basal ant species H. saltator and Di. quadriceps are able to mate and lay fertilized eggs. Both of these species retained the OD2-like domains in DSX, while all the more advanced ant species analyzed here have lost the OD2 domain, raising the possibility that changes in the ant dsx domain organization were possibly associated with the evolution of some crucial functions associated with eusocial life in ants, such as caste determination.
It is well known that the network comprising IIS and other pathways controls the development of different castes. These pathways integrate the different external stimuli such as nutrition and temperature, and ultimately initiate the different caste fates (queen and worker). However, the genetic mechanisms of developmental responses to IIS and other pathways are still unclear (Corona et al. 2016). In stag beetles, dsx interacts with nutrition-related pathways, such as the JH signaling pathway, to regulate sex-specific mandible growth (Gotoh et al. 2014). Recently, dsx was found to have sex- and morph-specific expression at different developmental stages in the ant C. obscurior (Klein et al. 2016). Similarly RNA-seq data revealed that dsx was highly differentially expressed across different tissues of honey bee nurses and foragers, suggesting that dsx is a possible candidate gene for the key regulator of caste-specific gene expression (Johnson and Jasper 2016). Taken together, these studies combined with our evolutionary analysis suggest that dsx may function as a “connector” in the caste determination cascade. dsx may be sensitive to hormones, and mediate the signal transduction from the initial input of nutrition or other environmental cues to effectors that control development of different traits. Finally, this connector model indicates that large evolutionary changes (e.g., caste differentiation) can be achieved by changes in gene regulation without the need to involve novel gene formation. However, it cannot be ruled out that other mechanisms may be responsible for the lack of OD2 domains in ants. Future functional experiments will be needed to address the impact of the lack of the OD2 domain in ants.
In conclusion, this study suggests a specific pattern in the evolution of dsx that correlates with the evolution of eusociality in ants. In the ant species displaying an early evolutionary state of eusociality, dsx had ancestral gene structure. In contrast, the loss of the OD2 domain and positive selection on dsx indicate an extraordinarily high evolutionary rate in the advanced ant species. Besides, there are some exclusive patterns of expression of Dmrt genes in two genetic caste determination ants. All these results suggest that Dmrt genes may be involved in the development of caste-specific morphological features. More functional studies are necessary to more fully understand the role of Dmrt genes in ants.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31601854 to L.-Y.J., 31672336 to J.-H.X., and 31572313 to D.-W.H.). The computing resource was supported by HPC Platform, Scientific Information Center, Institute of Zoology, CAS, China.
Literature Cited
- Altschul SF. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17):3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An WQ, Cho SY, Ishii H, Wensink PC.. 1996. Sex-specific and non-sex-specific oligomerization domains in both of the doublesex transcription factors from Drosophila melanogaster. Mol Cell Biol. 16(6):3106–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BS, Wolfner MF.. 1988. A molecular analysis of doublesex, a bifunctional gene that controls both male and female sexual-differentiation in Drosophila Melanogaster. Genes Dev. 2(4):477–489. [DOI] [PubMed] [Google Scholar]
- Bellefroid EJ, et al. 2013. Expanding roles for the evolutionarily conserved Dmrt sex transcriptional regulators during embryogenesis. Cell Mol Life Sci. 70(20):3829–3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourke AFG. 1988. Worker reproduction in the higher eusocial Hymenoptera. Q Rev Biol. 63:291–311. [Google Scholar]
- Buschinger A. 1968. Untersuchungen anHarpagoxenus sublævis Nyl. (Hymenoptera, formicidæ). Insectes Soc. 15(1):89–104. [Google Scholar]
- Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng DF, Zhang ZL, He XF, Liang GW.. 2013. Validation of reference genes in Solenopsis invicta in different developmental stages, castes and tissues. PLoS One 8(2):e57718.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Huang ZY, Zhang JZ.. 2007. Sex-specific splicing of the honeybee doublesex gene reveals 300 million years of evolution at the bottom of the insect sex-determination pathway. Genetics 177(3):1733–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Wensink PC.. 1997. DNA binding by the male and female doublesex proteins of Drosophila melanogaster. J Biol Chem. 272(6):3185–3189. [DOI] [PubMed] [Google Scholar]
- Cho S, Wensink PC.. 1998. Linkage between oligomerization and DNA binding in Drosophila doublesex proteins. Biochemistry 37(32):11301–11308. [DOI] [PubMed] [Google Scholar]
- Cho SY, Wensink PC.. 1996. Purification and physical properties of the male and female double sex proteins of Drosophila. Proc Natl Acad Sci U S A. 93(5):2043–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona M, Libbrecht R, Wheeler DE.. 2016. Molecular mechanisms of phenotypic plasticity in social insects. Curr Opin Insect Sci. 13:55–60. [DOI] [PubMed] [Google Scholar]
- Criscuolo A. 2011. morePhyML: improving the phylogenetic tree space exploration with PhyML 3. Mol Phylogenet Evol. 61(3):944–948. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27(8):1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan JP, et al. 2013. New insights into the genomic organization and splicing of the doublesex gene, a terminal regulator of sexual differentiation in the silkworm Bombyx mori. PLoS One 8(11):e79703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 1998. Profile hidden Markov models. Bioinformatics 14(9):755–763. [DOI] [PubMed] [Google Scholar]
- Erdman SE, Burtis KC.. 1993. The Drosophila doublesex proteins share a novel zinc finger related DNA binding domain. EMBO J. 12(2):527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman SE, Chen HJ, Burtis KC.. 1996. Functional and genetic characterization of the oligomerization and DNA binding properties of the Drosophila doublesex proteins. Genetics 144(4):1639–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1978. Cases in which parsimony or compatibility methods will be positively misleading. Syst Zool. 27(4):401–410. [Google Scholar]
- Fournier D, et al. 2005. Clonal reproduction by males and females in the little fire ant. Nature 435(7046):1230–1234. [DOI] [PubMed] [Google Scholar]
- Gadau J, et al. 2012. The genomic impact of 100 million years of social evolution in seven ant species. Trends Genet. 28(1):14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett-Engele CM, et al. 2002. intersex, a gene required for female sexual development in Drosophila, is expressed in both sexes and functions together with doublesex to regulate terminal differentiation. Development 129(20):4661–4675. [DOI] [PubMed] [Google Scholar]
- Geuverink E, Beukeboom LW.. 2014. Phylogenetic distribution and evolutionary dynamics of the sex determination genes doublesex and transformer in insects. Sex Dev. 8(1–3):38–49. [DOI] [PubMed] [Google Scholar]
- Goldman N, Yang ZH.. 1994. Codon-based model of nucleotide substitution for protein-coding DNA-sequences. Mol Biol Evol. 11:725–736. [DOI] [PubMed] [Google Scholar]
- Gotoh H, et al. 2014. Developmental link between sex and nutrition; doublesex regulates sex-specific mandible growth via juvenile hormone signaling in stag beetles. PLoS Genet. 10(1):e1004098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölldobler B, Wilson EO.. 1990. The ants. London: Harvard University Press. [Google Scholar]
- Hölldobler B, Wilson EO.. 2009. The superorganism: the beauty, elegance, and strangeness of insect societies. New York: W.W. Norton & Company. [Google Scholar]
- Helmkampf M, Cash E, Gadau J.. 2015. Evolution of the insect desaturase gene family with an emphasis on social Hymenoptera. Mol Biol Evol. 32(2):456–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BR, Jasper WC.. 2016. Complex patterns of differential expression in candidate master regulatory genes for social behavior in honey bees. Behav Ecol Sociobiol. 70(7):1033–1043. [Google Scholar]
- Kato Y, et al. 2008. Molecular cloning and sexually dimorphic expression of DM-domain genes in Daphnia magna. Genomics 91(1):94–101. [DOI] [PubMed] [Google Scholar]
- Kato Y, Kobayashi K, Watanabe H, Iguchi T.. 2011. Environmental sex determination in the branchiopod crustacean Daphnia magna: deep conservation of a Doublesex gene in the sex-determining pathway. PLoS Genet. 7(3):e1001345.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A, et al. 2016. Evolution of social insect polyphenism facilitated by the sex differentiation cascade. PLoS Genet. 12(3):e1005952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger MJB, Ross KG.. 2002. Identification of a major gene regulating complex social behavior. Science 295(5553):328–332. [DOI] [PubMed] [Google Scholar]
- Li YF, Rao XY, Mattox WW, Amos CI, Liu B.. 2015. RNA-seq analysis of differential splice junction usage and intron retentions by DEXSeq. PLoS One 10(9):e0136653.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livramento K, Freitas N, Máximo W, Zanetti R, Paiva L.. 2018. Gene expression profile analysis is directly affected by the selected reference gene: the case of leaf-cutting Atta sexdens. Insects 9(1):18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res. 43(Database issue):D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson CK, Zarkower D.. 2012. Sex and the singular DM domain: insights into sexual regulation, evolution and plasticity. Nat Rev Genet. 13(3):163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau CS, Bell CD.. 2013. Testing the museum versus cradle tropical biological diversity hypothesis: phylogeny, diversification, and ancestral biogeographic range evolution of the ants. Evolution 67(8):2240–2257. [DOI] [PubMed] [Google Scholar]
- Nagoshi RN, McKeown M, Burtis KC, Belote JM, Baker BS.. 1988. The control of alternative splicing at genes regulating sexual differentiation in D. melanogaster. Cell 53(2):229–236. [DOI] [PubMed] [Google Scholar]
- Ohkawara K, Nakayama M, Satoh A, Trindl A, Heinze J.. 2006. Clonal reproduction and genetic caste differences in a queen-polymorphic ant, Vollenhovia emeryi. Biol Lett. 2(3):359–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottolenghi C, Fellous M, Barbieri M, McElreavey K.. 2002. Novel paralogy relations among human chromosomes support a link between the phylogeny of doublesex-related genes and the evolution of sex determination. Genomics 79(3):333–343. [DOI] [PubMed] [Google Scholar]
- Peeters C, Liebig J, Holldobler B.. 2000. Sexual reproduction by both queens and workers in the ponerine ant Harpegnathos saltator. Insectes Soc. 47(4):325–332. [Google Scholar]
- Pomerantz AF, Hoy MA.. 2015. Expression analysis of Drosophila doublesex, transformer-2, intersex, fruitless-like, and vitellogenin homologs in the parahaploid predator Metaseiulus occidentalis (Chelicerata: Acari: Phytoseiidae). Exp Appl Acarol. 65(1):1–16. [DOI] [PubMed] [Google Scholar]
- Price DC, Egizi A, Fonseca DM.. 2015. The ubiquity and ancestry of insect doublesex. Sci Rep. 5:13068.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2000. R language definition. Vienna (Austria: ): R Foundation for Statistical Computing. [Google Scholar]
- Rideout EJ, Dornan AJ, Neville MC, Eadie S, Goodwin SF.. 2010. Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nat Neurosci. 13(4):458–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez L, Gorfinkiel N, Guerrero I.. 2001. Sex determination genes control the development of the Drosophila genital disc, modulating the response to Hedgehog, Wingless and Decapentaplegic signals. Development 128(7):1033–1043. [DOI] [PubMed] [Google Scholar]
- Scharlaken B, et al. 2008. Reference gene selection for insect expression studies using quantitative real-time PCR: the head of the honeybee, Apis mellifera, after a bacterial challenge. J Insect Sci. 8(33):1–10. [Google Scholar]
- Schwander T, Lo N, Beekman M, Oldroyd BP, Keller L.. 2010. Nature versus nurture in social insect caste differentiation. Trends Ecol Evol. 25(5):275–282. [DOI] [PubMed] [Google Scholar]
- Shirangi TR, Dufour HD, Williams TM, Carroll SB.. 2009. Rapid evolution of sex pheromone-producing enzyme expression in Drosophila. PLoS Biol. 7(8):e1000168.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota K, et al. 2013. Molecular cloning of doublesex genes of four Cladocera (water flea) species. BMC Genomics 14:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, et al. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 7(3):562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL.. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25(9):1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhulst EC, van de Zande L.. 2015. Double nexus—Doublesex is the connecting element in sex determination. Brief Funct Genomics. 14(6):396–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volff JN, Zarkower D, Bardwell VJ, Schartl M.. 2003. Evolutionary dynamics of the DM domain gene family in metazoans. J Mol Evol. 57:S241–S249. [DOI] [PubMed] [Google Scholar]
- Ward PS, Brady SG, Fisher BL, Schultz TR.. 2015. The evolution of myrmicine ants: phylogeny and biogeography of a hyperdiverse ant clade (Hymenoptera: Formicidae). Syst Entomol. 40(1):61–81. [Google Scholar]
- Wexler JR, Plachetzki DC, Kopp A.. 2014. Pan-metazoan phylogeny of the DMRT gene family: a framework for functional studies. Dev Genes Evol. 224(3):175–181. [DOI] [PubMed] [Google Scholar]
- Yang ZH. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang ZH, Nielsen R, Goldman N, Pedersen AMK. 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JZ, Nielsen R, Yang ZH.. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 22(12):2472–2479. [DOI] [PubMed] [Google Scholar]
- Zhang M, et al. 2014. Cloning, expression and cellular localization of the Doublesex gene in the water flea, Daphnia carinata, during different developmental stages. Gene 550(2):185–192. [DOI] [PubMed] [Google Scholar]
- Zhu LY, et al. 2000. Sexual dimorphism in diverse metazoans is regulated by a novel class of intertwined zinc fingers. Genes Dev. 14(14):1750–1764. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.