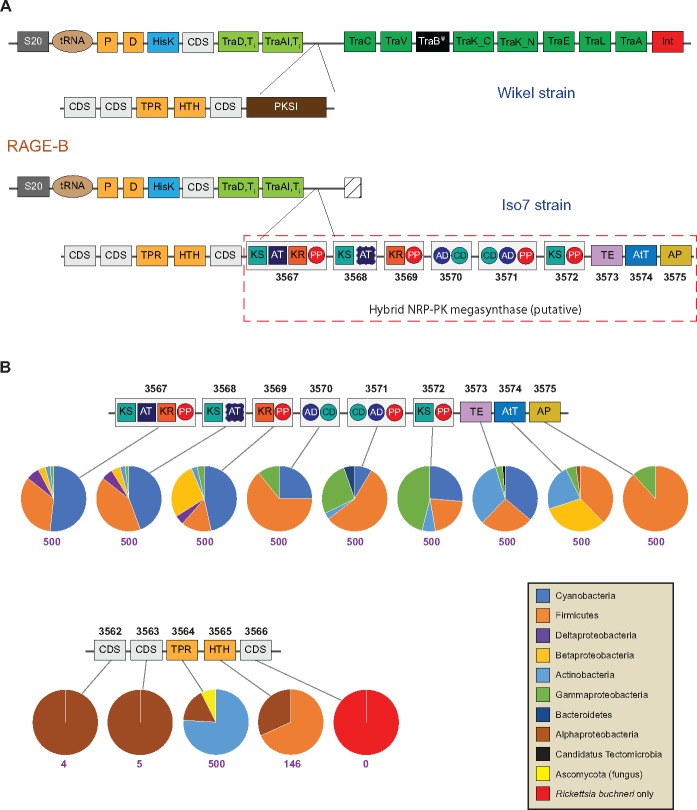
Fig. 4.
—RAGE-B harbors a multigene insertion that is distinctly nonalphaproteobacterial and strongly reminiscent of a hybrid NRPS/PKS cluster. (A) Comparison of gene order and functional annotation between RAGE-B variants from Rickettsia buchneri str. Wikel (top) and R. buchneri str. ISO7 (bottom). Functional symbols and color schemes generally follow figure 2. A red dotted line surrounds the nine genes that comprise the putative NRPS/PKS cluster in R. buchneri str. ISO7; the GenBank accession identifier for each protein is shown in black text below the corresponding cartoon representation (note: “KDO0” has been omitted from the start of each ID for brevity). Proteins in the putative cluster have been labeled with their predicted domains: KS, ketosynthase (teal); AT, acyltransferase (navy blue); KR, ketoreductase (orange); PP, phosphopantetheine binding (red oval); TE, thioesterase (lavender); AtT, acetyltransferase (blue); AP, aminopeptidase (gold). Proteins predicted to contain multiple domains are surrounded by a light gray rectangle. (B) Nonalphaproteobacterial origin for the 14 proteins of the RAGE-B multigene insertion. The putative NRPS/PKS cluster (top) and the remaining five genes of the RAGE-B insert (bottom) are shown in cartoon representation as in (A), along with pie charts showing the taxonomic distribution of their corresponding top BLASTP matches. Numbers below each pie chart (purple) indicate the total number of unique hits used to construct the distribution (maximum of 500).