Figure 7.
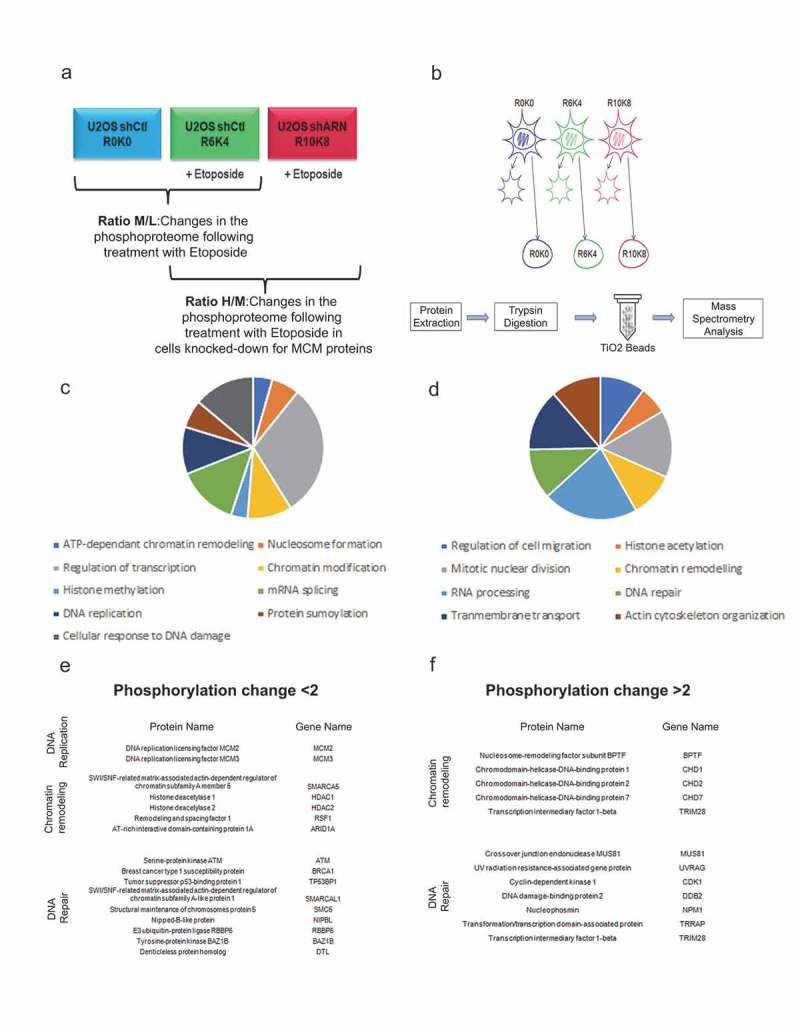
Effects of a decrease in the MCM complex on the phosphoproteome in response to double-strand breaks. U2OS cells infected with shRNAs against MCM2 or MCM3 were cultured in different SILAC media and treated with Etoposide, fractionated and then digested to purify the phosphopeptides. (a) The U2OS shControl cell line was cultured in light SILAC medium (R0K0) and untreated, or cultured in SILAC medium (R6K4) and treated for 1 hour with 10 μM Etoposide. U2OS cells infected with shRNAs against MCM2 or MCM3 were cultured in heavy SILAC medium (R10K8) and subsequently treated for 1 hour with 10 μM Etoposide. (b) The cells were fractionated to isolate the nuclear proteins, after which the proteins were extracted, digested with trypsin and purified on titanium dioxide beads before identification and quantification by mass spectrometry. (c,d) Proteins with a >2-fold increase (c) or >2-fold decrease (d) in phosphorylation were analyzed for gene ontology pathway enrichment using David 2.0. The pathways including at least four proteins and those with a p-value <0.01 for an increase or decrease in phosphorylation are presented. (e,f). Lists of proteins identified and known to play a role in chromatin remodeling, DNA repair or DNA replication mechanisms in response to DNA damage.
