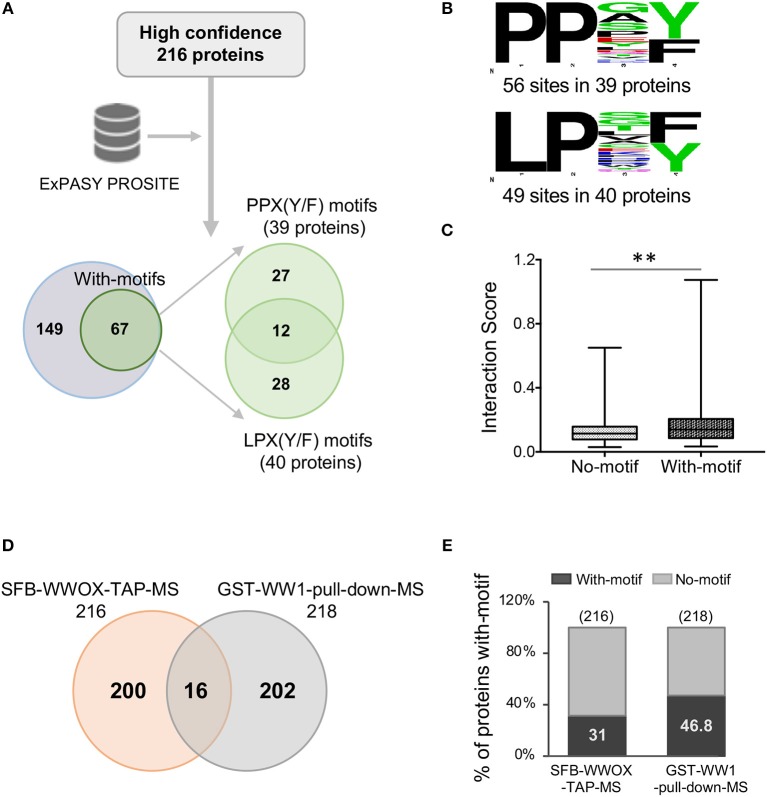
Figure 4.
WW domain binding motifs in WWOX interacting proteins. (A) ExPASY PROSITE analysis was performed on the list of 216 WWOX interactors (Supplementary File 2). Candidate WWOX WW1 domain binding motifs were identified in 31% (67 out of 216) proteins. Out of 67 proteins with putative WW1 binding motifs, 12 candidate WWOX binding interactors displayed both PPX(Y/F) and LPX(Y/F) motifs. (B) Amino acid sequence of candidate WWOX WW1 domain binding motifs. The probability of the residue at each position is proportional to the size of the letters. The image was generated using the Weblogo 3.4 program. Thirty-nine proteins displayed PPX(Y/F) motifs at 56 sites and 40 proteins displayed LPX(Y/F) motif at 49 sites. (C) Box and whisker plot showing the comparison of median interaction scores between proteins with- and without-motifs. Proteins with-motifs display higher interaction scores (mean interaction score = 0.1875) vs. proteins with No-motifs (mean interaction score = 0.1287). Statistical analysis was done using two-tailed unpaired Student's t-test, **p < 0.001. (D) Comparison of SFB-WWOX-TAP-MS data with GST-WW1-pull-down-MS data from Abu-Odeh (34). Only 16 common WWOX interactors were identified overlapping both datasets. (E) Bar graph showing a comparison of the percentage of proteins with WWOX binding motifs between the aforementioned datasets. Thirty-one percent of proteins from the SFB-WWOX-TAP-MS dataset (this study) displayed candidate WW1 binding motifs vs. 46.8% of proteins in the GST-WW1-pull-down-MS dataset (34).