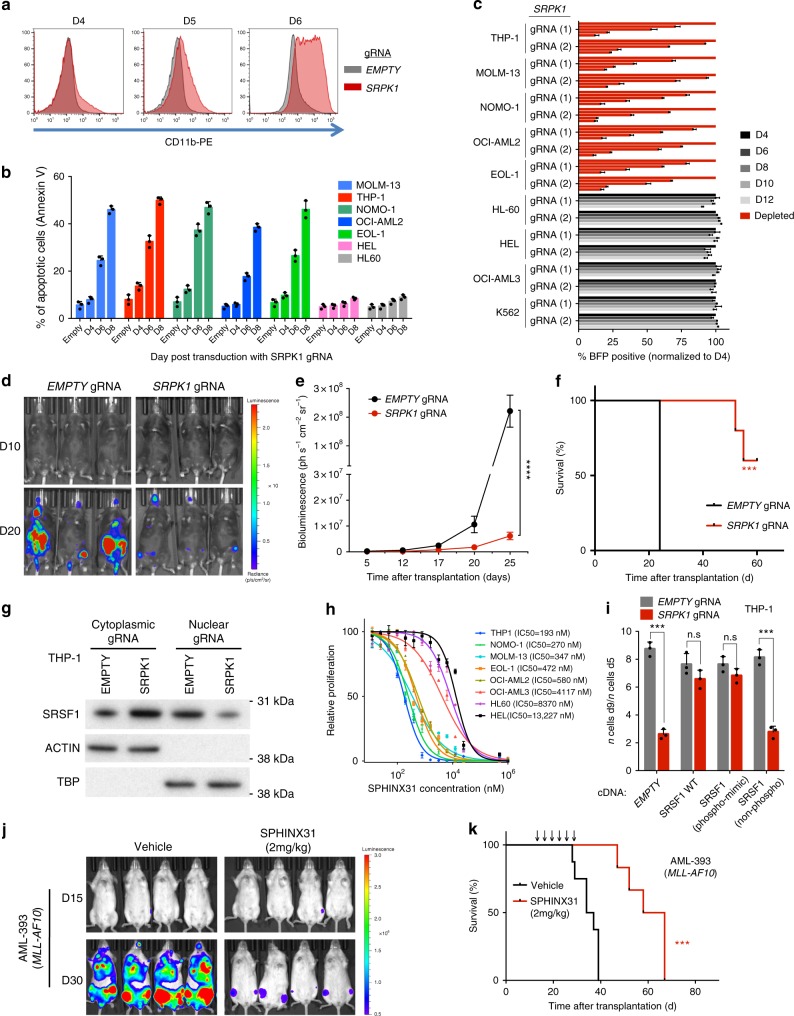
Fig. 1.
Genetic and pharmacological inhibition of SRPK1 inhibits growth and drives differentiation of human AML cells. a CD11b expression in MOLM-13 cells 4, 5, and 6 days after gRNA. b Increased apoptosis levels in AML cells driven by MLL-X fusion genes (MOLM13, THP1, NOMO-1, and OCI-AML2) or MLL-PTD (EOL-1) after dual gRNA targeting of SRPK1 (mean ± s.d., n = 3). c Competitive co-culture of lentiviral SRPK1 gRNA-transfected (BFP positive) vs untransfected AML cell lines normalized to %BFP on day 4 (mean ± s.d., n = 3). d Bioluminescence imaging of mice transplanted with MOLM-13-Cas9 cells transduced with luciferase-expressing lentiviral gRNAs. e Whole-body luminescence of mice depicted in (f) (n = 5). f Kaplan–Meier survival of MOLM-13-transplanted mice (n = 5). g Nuclear and cytoplasmic protein levels of SRSF1 after gRNA targeting of SRPK1 or empty in THP1 cells. h Dose-response curves of AML cell lines to the SRPK1 inhibitor SPHINX31 on day 6 post-treatment (mean ± s.d., n = 3), reveal that cell lines driven by MLL rearrangements are significantly more sensitive. i Proliferation of THP-1 cells transduced with gRNA targeting SRPK1 or EMPTY, and plasmids expressing a wild type (WT), a phosphomimic, a non-phosphorylatable version of SRSF1 or no cDNA (EMPTY) (mean ± s.d., n = 3). j Bioluminescence imaging of luciferase-expressing, MLL-X-driven, AML PDX models, treated with 2 mg/kg SPHINX31 (n = 6). Extended data in Supplementary Figure 4. k Kaplan–Meier survival of MLL-X-driven, AML PDX models, treated with 2 mg/kg SPHINX31 at indicated times (arrows) (n = 6). ***p < 0.001 (t-test). ****p < 0.0001 (t-test). d, day; Log-rank (Mantel–Cox) test was used for survival comparisons