FIG 3.
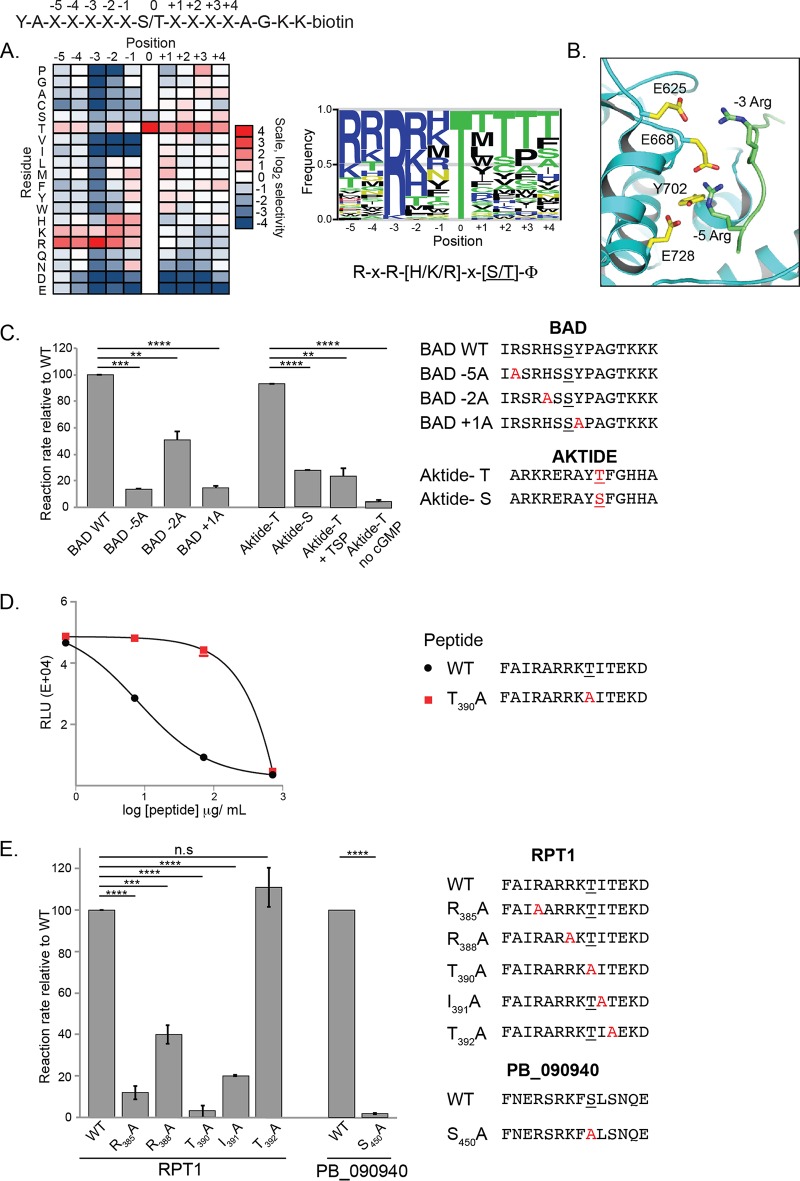
PfPKG has a consensus phosphorylation site distinct from that of mammalian PKG. (A) A combinatorial peptide library consisting of 182 peptide mixtures with the general sequence shown at the top of the panel was used as the substrate in PfPKG enzymatic assays. The heat map shows the relative levels of phosphorylation of peptide mixtures having the indicated amino acid present at the indicated position. The sequence logo is scaled such that the height of the letter is proportional to the level of phosphorylation of the corresponding amino acid residue at the indicated position within the peptide. The logo was prepared using enoLOGOS (53). Results shown are the average from two separate determinations. (B) The X-ray crystal structure of PfPKG (PDB accession number 5DYK) was overlaid with that of an AKT-substrate peptide complex (PDB accession number 1O6K) using PyMOL. The substrate peptide (green) is modeled bound to PfPKG (cyan; the indicated interacting residues are shown in yellow). For clarity, AKT is not shown. (C) The consensus phosphorylation site of PfPKG was confirmed by determining the reaction rate of PfPKG using individual peptides as substrates. Peptides with amino acid replacements at key positions were used as substrates in PfPKG activity assays. The presumed phosphoacceptor site in each peptide is underlined. Reaction rates for each peptide were determined relative to the rate of the wild type (WT). PfPKG has a strong preference for amino acids at the −5 and +1 positions. PfPKG prefers Thr (AKTIDE-T versus AKTIDE-S) as the phosphoacceptor, a basic residue at the −5 position (BAD wild type versus BAD-5A), and a hydrophobic residue at the +1 position (BAD wild type versus BAD + 1A) relative to the phosphorylation site. It has weaker preference for peptides containing basic residues at the −2 position (BAD wild type versus BAD-2A). Results are the average of two experiments, each done in triplicate, ± standard deviations. (D) The domain of RPT1 that contacts PfPKG contains a phosphorylation site for PfPKG. Split-luciferase complementation assays between Nfluc-PfPKG E706A and Cfluc-RPT1114–420 were performed in the presence of peptides carrying either the wild-type (WT) or a mutant (T390A) consensus phosphorylation site from RPT1114–420. Wild-type peptide, but not the mutant, competed with RPT1 for interaction with PfPKG. Results are the average of three experiments, each performed in triplicate. (E) PfPKG efficiently phosphorylates peptides matching its consensus sequence present in two possible substrate proteins, RPT1 and PB_090940. Peptides with amino acid replacements at key positions were used as substrates in PfPKG activity assays. The presumed phosphoacceptor site in each peptide is underlined. Reaction rates for each peptide were determined relative to the rate of the wild type. Results are the average of three experiments, each performed using two or three replicates. Data were analyzed using an unpaired t test (**, P < 0.001; ***, P = 0.0001; ****, P < 0.0001; ns, not significant).