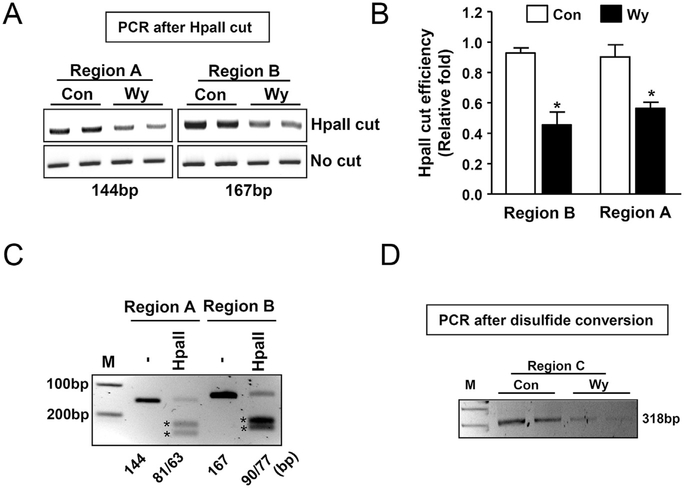
Fig. 3. Wy-14,643, a PPAR𝛂 agonist, decreases CpG methylation in the Gadd45b promoter region.
(A) To examine the cytosine methylation pattern in the selected CpG sequences, mice were fed with Wy-14,643 (0.1%, w/w) for 1 day and isolated liver genomic DNA was incubated with enzyme HpaII, a methylation sensitive restriction enzyme for epigenetic and cutting regions were amplified using conventional PCR reaction. (B) HpaII cut efficiency represented as a densitometry from (A). (C) To verify the correct PCR products from (A), PCR products were cut with the HpaII enzyme. The unmethylated PCR products were cut into two fragments to verify the correct sizes. Stars indicate the cutting sizes. (D) To demonstrate the epigenetic change of the Gadd45b promoter by Wy-14,643, the disulfide conversion method was applied. Unmethylated cytosine was delaminated to form uracil and samples were subjected to conventional PCR analysis with specifically designed primers. Con, control; Wy, Wy-14,643 (0.1%, w/w); *p < 0.05.