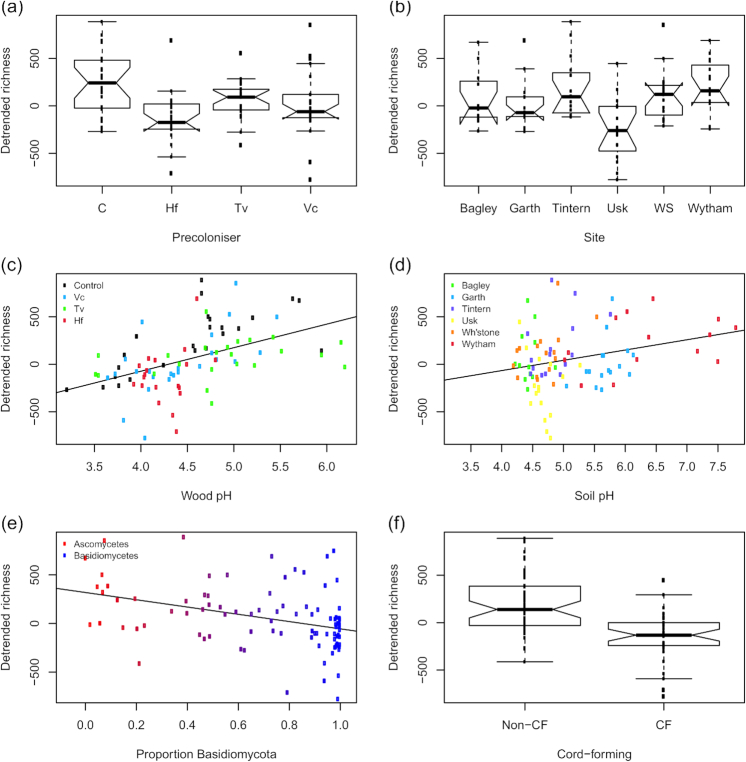
Figure 4.
Overall bacterial OTU richness in fungus-colonised wood disks. Residuals from a model to correct for sequencing depth, broken down by (a) pre-coloniser identity; (b) site of origin; (c) wood pH (coloured by pre-coloniser); (d) soil pH (coloured by site); (e) relative proportions of ascomycete and basidiomycete reads and (f) whether the dominant fungal OTU belonged to a cord-forming genus. Individual data points are overlaid on boxplots. The y-scale relates to the number of OTUs relative to that predicted by sequencing depth alone, i.e. negative values denote samples with lower diversity than predicted by the null model. A simple regression line is overlaid on the scatterplots. Notches on boxplots represent 95% confidence intervals; where these extend beyond the quartiles, ‘hinges’ appear on the plot. Abbreviations are control (C), Vulleminia comedens (Vc), Trametes versicolor (Tv), Hypholoma fasciculare (Hf), Whitestone (WS), cord-former (CF).