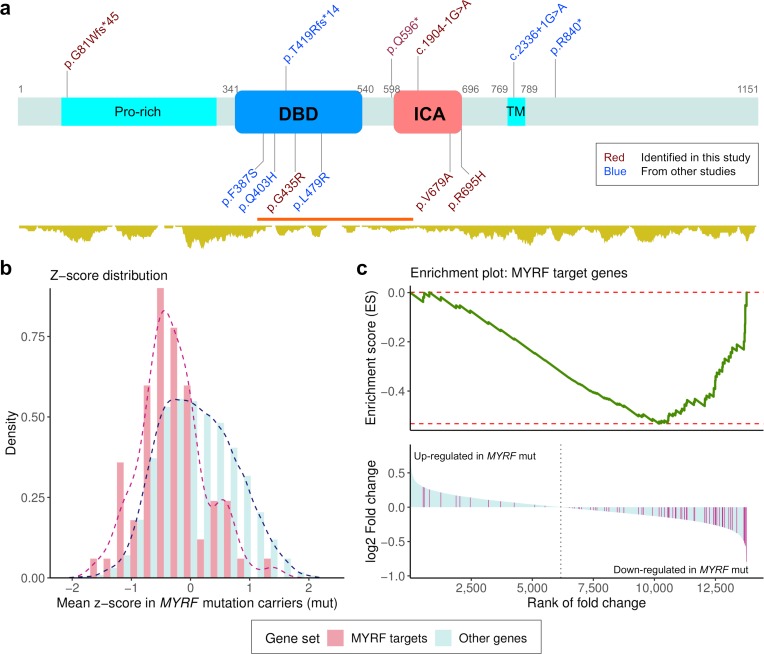
Fig 1. De novo coding variants in MYRF and their functional impact on transcriptome.
(a) Schematic diagram of the MYRF protein structure. DBD: DNA binding domain; ICA: Intramolecular chaperone auto-processing domain; Pro-rich: proline-rich region; TM: transmembrane helix. The position of DBD and ICA were based on the annotation from InterPro, and Pro-Rich and TM were from SwissProt. The coordinates are given with respect to the canonical isoform (1151 amino acids). The relative position of 12 de novo coding variants are displayed, including 6 discovered in the current study (shown in red), and five from published studies of congenital heart disease (CHD) [29, 30] (shown in blue). LGD variants are shown on top of the protein; and missense variants are on the other side. Shown below the protein structure is the density of missense variants in gnomAD (http://gnomad.broadinstitute.org/). A missense constraint region [37] is highlighted in red (observed/expected number of missense variants = 0.31) (b) Z-score for each gene is the standardized expression level across samples. Mean Z-scores of MYRF target genes in three MYRF variant carriers were shifted to the lower end as compared with other genes. (c) Gene-set enrichment analysis (GESA) was applied to genes ranked by the estimated fold change of expression level comparing MYRF variant carriers with other cases. The MYRF target genes tend to have lower ranks and majority of them were down-regulated in MYRF variant carriers (NES = -2.10, P<5.0E-4).