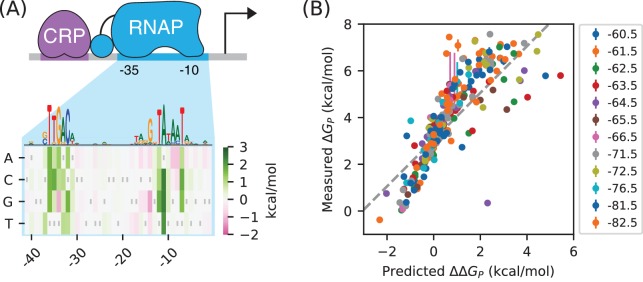
Figure 6. RNAP-DNA binding energy cannot be accurately predicted from sequence.
(A) The PSAM for RNAP-DNA binding inferred by Kinney et al. (2010). This model assumes that the DNA base pair at each position in the RNAP binding site contributes independently to . Shown are the values assigned by this model to mutations away from the lac* RNAP site. The sequence of the lac* RNAP site is indicated by gray vertical bars; see also Appendix 1—figure 1. A sequence logo representation for this PSAM is provided for reference. (B) PSAM predictions plotted against the values inferred by fitting the allelic manifolds in Figure 5C. Error bars on these measurements represent 68% confidence intervals. Note that measured values are absolute, whereas the predictions of the PSAM are relative to the lac* RNAP site, which thus corresponds to kcal/mol. The dashed line, provided for reference, has slope 1 and passes through this lac* data point.