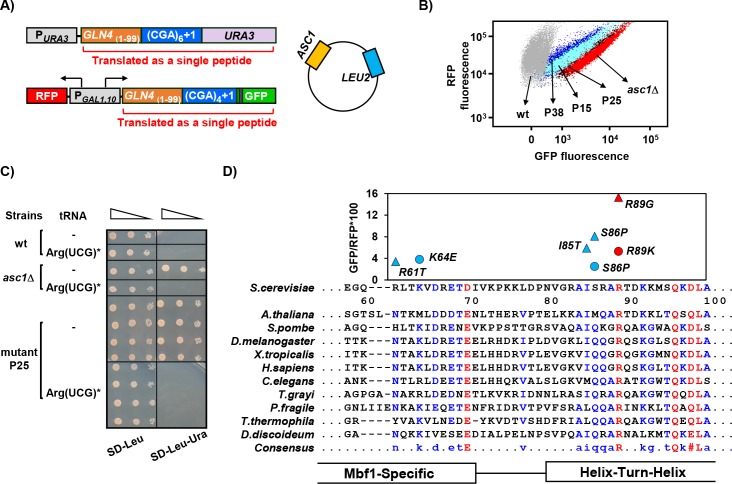
Figure 1. MBF1 (Multi-protein Bridging Factor 1) prevents frameshifting at CGA codon repeats.
(A) Schematic of selection for mutants that frameshift at CGA codon repeats. The indicated CGA codon repeats plus one extra nucleotide were inserted upstream of the URA3 and GFP coding region (with an upstream HA tag shown in purple), resulting in an Ura- GFP- parent strain. Additional copies of the ASC1 gene were introduced on a LEU2 plasmid to avoid recessive mutations in the native ASC1 gene. To obtain mutants with increased frameshifting efficiency, Ura+ mutants were selected and screened for increased GFP/RFP. (B) Expression of GLN4(1-99)-(CGA)4+1-GFP is increased in the MATa Ura+ mutants from this selection. Flow cytometry scatter plot showing GFP fluorescence versus RFP fluorescence for three mutants from this selection (P15: mbf1-R89K, light blue; P25: mbf1Δ125–151, black; P38: mbf1-K64E, dark blue), for the asc1Δ mutant (red) and for the wild-type parent strain (gray). (C) Expression of the non-native tRNAArg(UCG)* suppressed the Ura+ phenotype of mutant P25 at 30°C. Serial dilutions of the indicated strains with empty vector or expressing the mutant tRNAArg(UCG)* were grown on the indicated media. (D) Mutations in the MBF1 mutants map in conserved amino acids in both the MBF1-specific domain and the Helix-Turn-Helix (HTH) domain of Mbf1 protein. Alignment of yeast Mbf1 amino acids 60–100 with other eukaryotic species is shown (full alignment see Figure 1—figure supplement 3A). GFP/RFP of frameshifted (CGA)4+1 reporter is shown for mutants obtained from MATa (circles) and MATα (triangles) strains, with the color of markers corresponding to the consensus level of this residue (Blue: 50–90%, Red: >90%), however the conserved residue for R61 is N, and for S86 is Q, with all others identical to yeast.