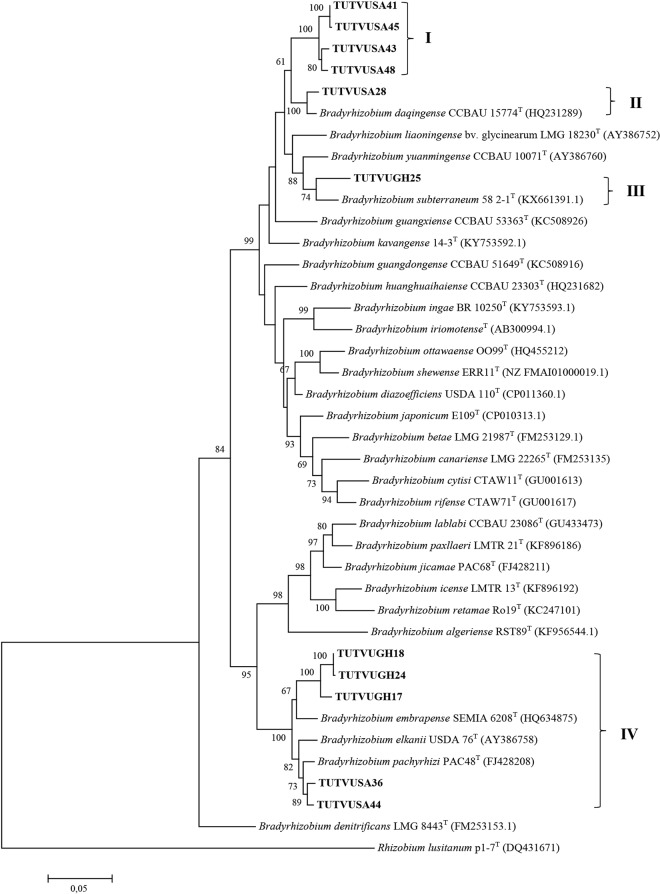
Figure 3.
Maximum likelihood phylogeny of cowpea nodulating rhizobia from Ghana and South Africa based on concatenated sequences of atpD-glnII-gyrB-rpoB genes. The evolutionary history was inferred by using the Maximum Likelihood method based on the Kimura 2-parameter model50. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 40 nucleotide sequences. We used only those strains which were present in all test gene phylogenies. Codon positions included were 1st + 2nd + 3rd + Noncoding. All positions containing gaps and missing data were eliminated. There were a total of 1523 positions in the final dataset. Evolutionary analyses were conducted in MEGA749.