Figure 10.
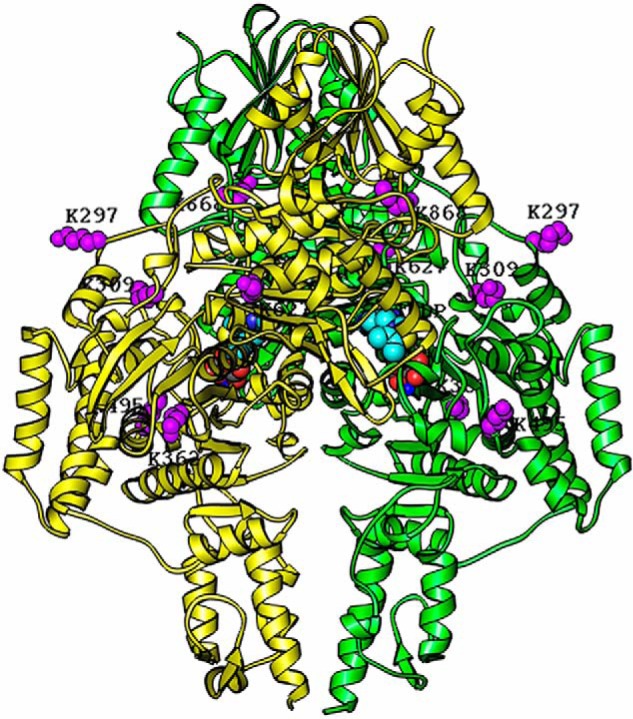
Homology modeling of the hE1o dimer structure with lysine residues involved in inter-component cross-linking highlighted in purple. The structure of the hE1o dimer was modeled based on amino acid sequence homology with the E. coli E1o (PDB code 2JGD (35)) and M. smegmatis E1o (PDB code 2YIC (39)) whose X-ray structures are available. The hE1o homodimer is colored in yellow and green. The identified inter-component cross-linked lysine residues (Lys297, Lys309, Lys362, Lys495, Lys627, and Lys868) are highlighted in purple and were positioned by sequence alignment of the hE1o with E. coli E1o. The identified lysine residues and ThDP cofactors are shown as space-filling representations. The program RIBBONS (6) was used to create this figure.
