Figure 5.
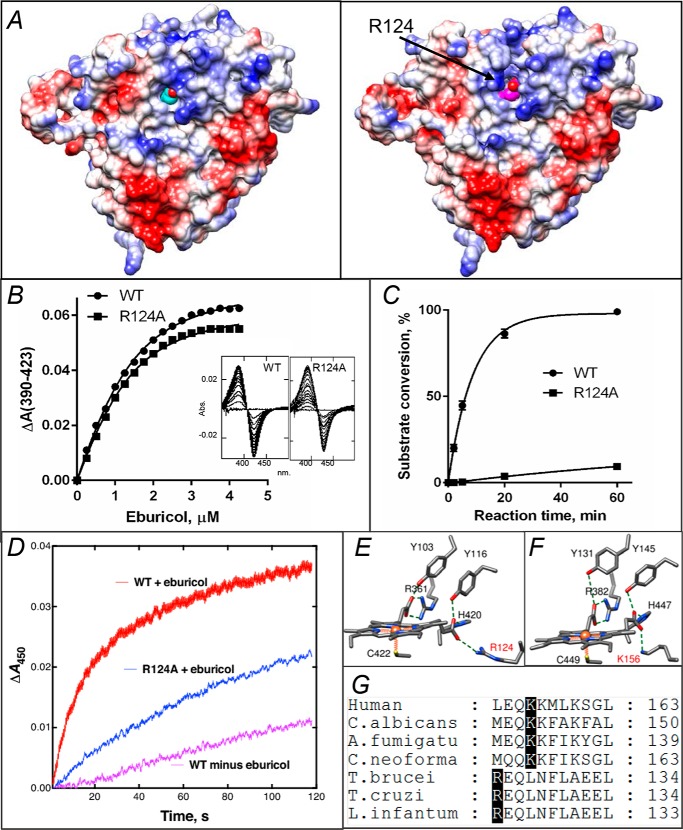
In protozoan CYP51, Arg-124 must be involved into the electron transfer. A, proximal surface of the ligand-free (left panel) and substrate-bound (right panel) I105F T. cruzi CYP51 molecules colored by electrostatic potential (red for negative and blue for positive charge). The heme is seen in cyan and magenta, respectively. In the substrate-bound structure repositioning of helix C and exposure of the guanidino group of Arg-124 (marked with an arrow) adds positive charge to the surface, whereas in the substrate-free structure the guanidino group of Arg- 124 forms the H-bond with the heme ring D propionate. B, spectral response of the R124A mutant to the substrate binding, 2 μm P450; Kd = 0.6 μm; the high-spin form content 36% (fit to Morrison equation nonlinear regression). The response of the WT protein (Kd 0.5 μm, 39% high-spin) is shown as a comparison. C, time course of substrate conversion by T. cruzi CYP51, WT, and the R124A mutant. The reaction mixture contained 1 μm P450, 2 μm T. brucei CPR, and 50 μm eburicol. D, enzymatic reduction. Rates of reduction of (ferric) T. cruzi WT and R124A were measured under anaerobic conditions in the presence of CO at 23 °C, using final concentrations of 1 μm P450, 2 μm CPR, 150 μm DLPC, and 5 μm eburicol and estimated by global fitting of the 380–520-nm data points (from >4 individual reductions, ± S.D.) to first-order plots in the OLIS software: WT enzyme, 1.56 ± 0.24 min−1; WT minus substrate, 0.20 ± 0.02 min−1; R124A enzyme (with substrate), 0.65 ± 0.02 min−1. E, heme support in protozoan CYP51, ligand-free T. brucei, PDB code 3G1Q. F, heme support in VFV-bound human CYP51, PDB code 4UHI. Arg-124 and Lys-156 are labeled in red. G, fragment of CYP51 sequence alignment showing the Arg and Lys residues that form the salt bridge with the heme.