Figure 2.
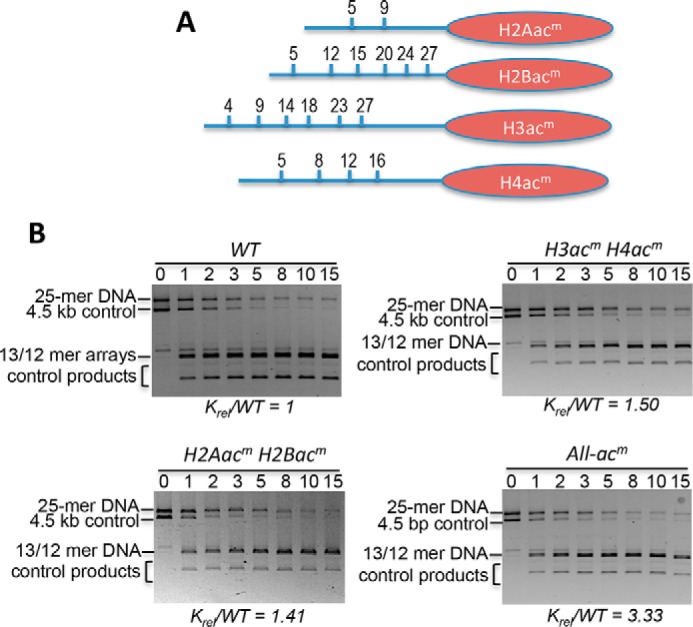
Acetylation mimics in all core histone tail domains increase accessibility of central target nucleosome linker DNA within a condensed 25-mer array. A, location of Lys → Gln substitutions as acetylation mimics (acm) in each of the core histone N-terminal tail domains (blue lines). B, representative SDS-agarose gels showing the DraIII digestion time course for 25-mer arrays with TNuc containing the indicated modified histones. Digestions were carried out for the times indicated above the lanes, and then products were separated on 0.7% SDS-agarose gels and stained with ethidium bromide. Gels were quantified, and rates of digestions were determined as described under “Experimental procedures.” The relative rate of digestion for each construct, normalized to the WT TNuc, is shown below the gel. See also Table 1. Bands corresponding to the 5.3-kb 25-mer array template (array) and the 4.5-kb naked control DNA (DNA) are indicated.
