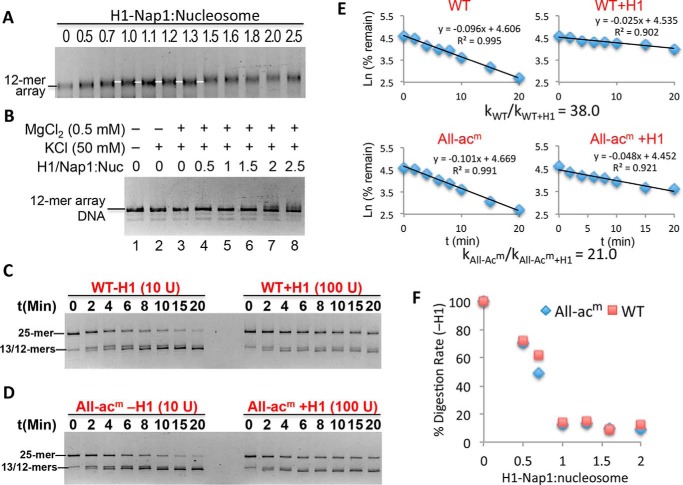
Figure 3.
Stoichiometric binding of H1 drastically decreases linker DNA accessibility. A, deposition of H1 onto nucleosome arrays via Nap1. Arrays were incubated with increasing amounts of H1–Nap1 complex, and binding was analyzed on a native agarose gel stained with ethidium bromide. The white lines indicate a species with a distinct mobility that arises over a range of H1–Nap1:nucleosome ratios (numbers above the lanes) consistent with one H1 bound to each nucleosome (white lines). B, self-association assay showing H1-bound arrays are soluble in 0.5 mm Mg2+. Nucleosome arrays were incubated as in A in H1 binding buffer (50 mm KCl) and 0.5 mm Mg2+ and centrifuged, and the DNA content of the supernatant was assessed on SDS-agarose gels. C, stoichiometric binding of H1 drastically reduces accessibility of TNuc linker DNA. 25-mer arrays containing a WT nucleosome at the central (TNuc) position were digested with DraIII either in the absence or presence of a 1.3:1 ratio of H1–Nap1:nucleosome. Digestions were carried out in buffer containing 0.5 mm Mg2+. Note that chromatin in the absence or presence of H1 was digested with either 10 or 100 units of DraIII, respectively. Digestion rates were determined as described, and relative rates were calculated and adjusted for amount of enzyme in each reaction. D, as in C except that the central nucleosome contained histones with acetylation mimics in all four histone proteins (All-acm). E, data from C and D were plotted as described in the text. The relative digestion rates in the absence/presence of H1 for this experiment are shown. See also Table 1. F, rates of DraIII digestion were determined for WT and All-acm arrays in the absence or presence of increasing H1–Nap1. Plotted is the rate of digestion normalized to the rate in the absence of H1 (100%).