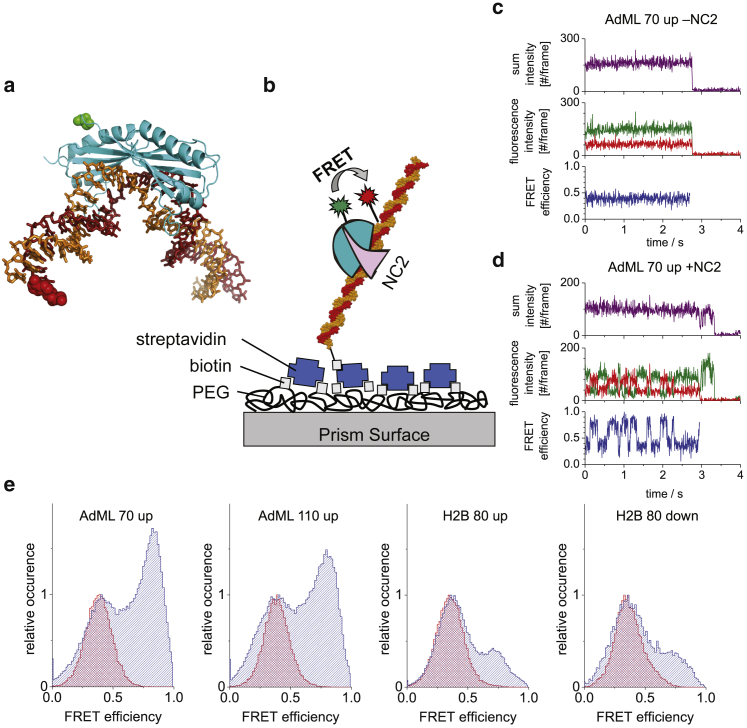
Figure 2.
SpFRET data of TBP-DNA in the absence and presence of NC2. (a) The crystal structure of TBP-DNA complex (Protein Data Bank: 1RM1) is shown. The labeling positions of the TBP (green) and DNA (red) are shown as spheres. For display purposes, the DNA has been extended downstream. (b) A scheme of the TBP-NC2-DNA complex immobilized to a functionalized, PEGylated glass surface over a streptavidin-biotin linkage is shown. TBP is specifically labeled with a donor fluorophore, and the DNA is specifically labeled with the acceptor fluorophore. The sample is excited by the evanescent wave generated at the prism/buffer interface by total internal reflection. (c and d) Single-molecule FRET trajectories from TBP bound to 70-bp DNA containing the AdML TATA box are plotted in the (c) absence and (d) presence of NC2 (purple: the scaled total intensity, , where γ is the detection correction factor; green: donor intensity; red: acceptor intensity; and blue: FRET efficiency). In the absence of NC2, a stable conformation of the TBP-DNA complex is observed, resulting in a constant FRET efficiency of ∼0.40. After the addition of NC2, the FRET efficiency fluctuates between different states. (e) Histograms of the frame-by-frame FRET efficiencies from the complete data set of the four different constructs measured are shown. Red histograms represent before the addition of NC2, whereas blue histograms represent after the addition of NC2.