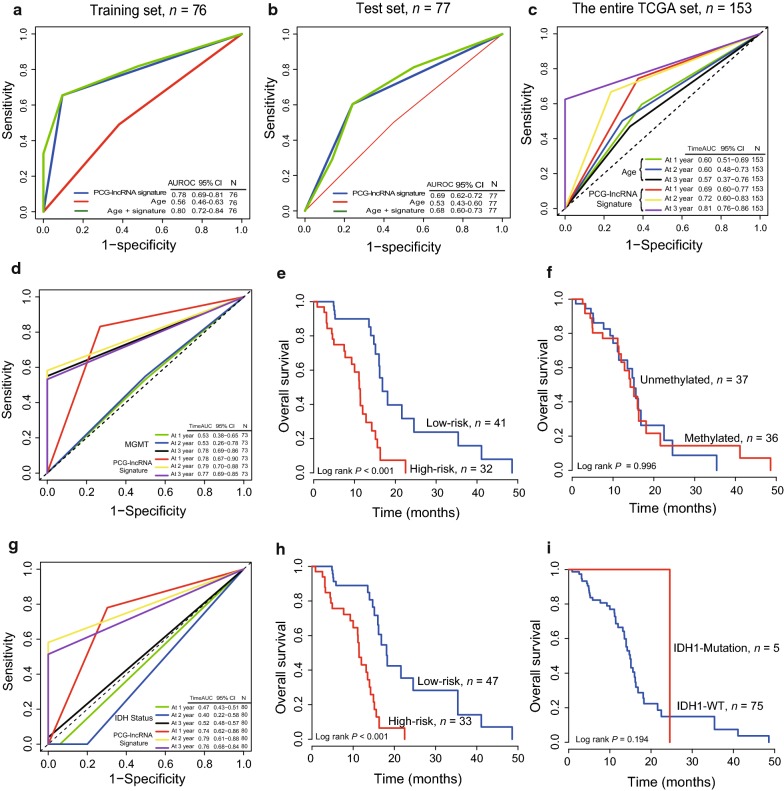
Fig. 3.
ROC analyses for comparison of the predictive power of the PCG-lncRNA signature with that of age in the training (a), test dataset (b) and TimeROC analysis in the entire set (c). TimeROC analysis for comparison of the survival predictive power of the PCG-lncRNA signature with that of MGMT promoter methylation status (d) and IDH1 mutation (g). Kaplan–Meier survival curves found the PCG-lncRNA signature (e, h) could classify patients with known MGMT promoter (f) and IDH1 mutation status (i) into high- and low-risk groups by the PCG-lncRNA signature in each corresponding TCGA dataset. Kaplan–Meier survival curves found the MGMT and IDH1/2 could not group patients into high- and low-risk groups