Abstract
A genetic predisposition to higher waist-to-hip ratio adjusted for BMI (WHRadjBMI), a measure of body fat distribution, associates with increased risk for type 2 diabetes. We conducted an exome-wide association study of coding variation in UK Biobank (405,569 individuals) to identify variants that lower WHRadjBMI and protect against type 2 diabetes. We identified four variants in the gene ACVR1C (encoding the activin receptor-like kinase 7 receptor expressed on adipocytes and pancreatic β-cells), which independently associated with reduced WHRadjBMI: Asn150His (−0.09 SD, P = 3.4 × 10−17), Ile195Thr (−0.15 SD, P = 1.0 × 10−9), Ile482Val (−0.019 SD, P = 1.6 × 10−5), and rs72927479 (−0.035 SD, P = 2.6 × 10−12). Carriers of these variants exhibited reduced percent abdominal fat in DEXA imaging. Pooling across all four variants, a 0.2 SD decrease in WHRadjBMI through ACVR1C was associated with a 30% lower risk of type 2 diabetes (odds ratio [OR] 0.70, 95% CI 0.63, 0.77; P = 5.6 × 10−13). In an analysis of exome sequences from 55,516 individuals, carriers of predicted damaging variants in ACVR1C were at 54% lower risk of type 2 diabetes (OR 0.46, 95% CI 0.27, 0.81; P = 0.006). These findings indicate that variants predicted to lead to loss of ACVR1C gene function influence body fat distribution and protect from type 2 diabetes.
Introduction
Discovery of genetic variants that protect against disease can identify novel mechanisms of disease and novel therapeutic targets (1). For example, the discovery of low-frequency coding variants in PCSK9, ANGPTL3, and APOC3 that lower blood lipid levels and protect against coronary artery disease catalyzed the development of novel therapeutics for coronary artery disease (2–10). PCSK9 inhibitors are now approved for treatment of coronary artery disease (4), while inhibitors of ANGPTL3 (7) and APOC3 (11) are in clinical development.
Body fat distribution strongly influences the development of type 2 diabetes (12–14). In a Mendelian randomization study of 296,291 individuals, we previously found that a genetic predisposition to increased abdominal fat distribution was associated with elevated triglyceride levels, elevated blood pressure, and an increased risk of coronary artery disease, independent of overall adiposity (12). Furthermore, a genetic predisposition to increased abdominal fat distribution was strongly associated with the development of type 2 diabetes. For each 1 SD genetic increase in waist-to-hip ratio adjusted for BMI (WHRadjBMI) (a measure of body fat distribution), risk of type 2 diabetes increased by 77% (12). These findings were replicated in a separate Mendelian randomization study (14).
These results suggest the hypothesis that genetic variants that influence body fat distribution may also influence the risk of type 2 diabetes (12). Here, we test this hypothesis by analyzing genetic variation in more than 400,000 individuals in UK Biobank to identify novel genetic variants that lower WHRadjBMI and protect against type 2 diabetes. Below, we demonstrate that variants predicted to lead to loss of function of the gene ACVR1C, which encodes the activin receptor-like kinase 7 (ALK7), influence body fat distribution and protect against type 2 diabetes.
Research Design and Methods
Study Design
In our discovery analysis, we analyzed the association of 614,042 coding variants with WHRadjBMI in 405,569 individuals in UK Biobank. Coding variants were defined as missense variants or variants predicted to result in loss of function of the protein: 1) nonsense mutations that resulted in early termination of a protein, 2) frameshift mutations due to insertions or deletions of DNA, or 2) splice-site mutations that result in an incorrectly spliced protein. Only variants imputed with a quality score (info score) >0.3 were analyzed. Threshold for significance was defined as P < 5 × 10−8 (genome-wide significance), and analysis was performed using PLINK2 software (15). We reported novel variants as those located more than one megabase away from previously identified loci in the Genetic Investigation of ANthropometric Traits (GIANT) Consortium (16). We reported variants located outside of the MHC locus separately from those within the MHC locus, as variants within the MHC locus typically tag HLA risk alleles and are thus associated with phenotypes due to linkage disequilibrium with HLA alleles (17). We attempted to replicate the association of novel variants with WHRadjBMI using data from the GIANT Consortium, when the variant was available in the GIANT Consortium (16).
Upon identification of variants in ACVR1C as significantly associated with WHRadjBMI, a conditional analysis was conducted to identify additional variants significantly associated (P < 0.0001) with WHRadjBMI within the ACVR1C locus (±250 kb of the lead variant rs55920843). To replicate observed associations of ACVR1C variants with WHRadjBMI, we examined whether carriers of these variants had reduced WHRadjBMI in a meta-analysis of the GIANT Consortium and two independent cohorts (Atherosclerosis Risk in Communities [ARIC] and Framingham Heart Study). We also examined whether these variants were associated with direct imaging-based measurements of abdominal fat in 4,215 participants who underwent DEXA imaging in UK Biobank.
To test whether identified ACVR1C variants were also associated with risk of type 2 diabetes, we pooled data from the DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium (ExTexT2D exome chip analysis [18]) with UK Biobank. To test whether variation leading to loss of ACVR1C function protects against type 2 diabetes, we analyzed the sequences of the nine exons of ACVR1C in 55,516 participants (31,672 from the Myocardial Infarction Genetics Consortium [MIGen] [19,20], 5,388 from the ARIC study, and 18,456 from the Type 2 Diabetes Genetic Exploration by Next-generation sequencing in multi-Ethnic Samples [T2D-GENES] Consortium [21]) and examined whether predicted damaging variants in the gene associate with risk of type 2 diabetes.
A phenome-wide association study of ACVR1C in UK Biobank was performed using an ACVR1C gene risk score (22). Definitions for 31 different diseases analyzed in the phenome-wide association study are provided (Supplementary Table 1). Three metabolic traits available in UK Biobank (urinary albumin-to-creatinine ratio, systolic blood pressure, and diastolic blood pressure) were also analyzed. A P value of 0.001 (0.05/34) was used for significance in this analysis.
Data Sources
For the analysis of WHRadjBMI, individual-level data from 405,569 unrelated individuals from the UK Biobank (335,660 individuals of European ancestry and 69,909 individuals of non-European ancestry) were analyzed. UK Biobank received ethical approval from the Research Ethics Committee (reference number 11/NW/0382). Analysis of UK Biobank was approved by the Partners Health Care Institutional Review Board (protocol 2013P001840). Informed consent was obtained from all participants by UK Biobank. For replication, data for WHRadjBMI from the GIANT Consortium (in which the Ile482Val variant was available) were pooled with data from the ARIC and Framingham Heart Study data sets (in which Asn150His, Ile195Thr, and rs72927479 variants were available). The GIANT Consortium consisted of 224,459 participants (210,088 of European ancestry and 14,371 of non-European ancestry) genotyped using the MetaboChip (16). The ARIC study is a community-based study of 15,792 white and black participants, aged 45 to 64 years (23). The ARIC data set consisted of 10,122 individuals (8,015 of European ancestry and 2,107 of non-European ancestry) who were genotyped and imputed, as previously described (24). For 5,388 participants, exome sequences were also available for analysis. In the Framingham Heart Study, a community-based study of 10,092 individuals of predominantly European ancestry, genotyped data were available from 6,073 individuals of European ancestry.
For the analysis of type 2 diabetes, estimates from UK Biobank were pooled using inverse variance–weighted fixed-effects meta-analysis with estimates from the DIAGRAM ExTexT2D exome chip analysis of 452,244 participants (81,412 case subjects with diabetes and 370,832 control subjects) (18). In UK Biobank, type 2 diabetes was defined as 1) self-report of type 2 diabetes, followed by a verbal interview with a trained nurse to confirm the diagnosis, or 2) hospitalization for ICD code E11. Because the ExTexT2D analysis included 120,286 participants from UK Biobank (18), these individuals were excluded from the analysis of type 2 diabetes in UK Biobank to prevent analysis of overlapping samples.
Sequence data for ACVR1C were extracted from exome sequencing performed in MIGen as previously described (19,20). The Burrows–Wheeler aligner algorithm was used to align reads from participants to the reference genome (hg19). The GATK HaplotypeCaller was used to jointly call variants. Metrics including variant quality score recalibration, quality over depth, and strand bias were then used to filter variants. The Jackson Heart Study (JHS) was excluded from analysis of MIGen, as it was included in the T2D-GENES Consortium. Exome sequences from 5,388 participants in ARIC were analyzed as previously described (25). Phenotype and genotype data were retrieved from the National Center for Biotechnology Information dbGaP server (accession phs000090.v3.p1 and phs000572.v6.p4). Exome sequencing was performed in the T2D-GENES Consortium as previously described (21). To analyze exome sequences from the T2D-GENES Consortium, the online Genetic Association Interactive Tool in the Type 2 Diabetes Knowledge Portal was used (21).
Studies included in MIGen were 1) the Italian Atherosclerosis, Thrombosis, and Vascular Biology (ATVB) study (dbGaP study accession phs000814.v1.p1); 2) the Exome Sequencing Project Early-Onset Myocardial Infarction (ESP-EOMI) study (9); 3) a nested case-control cohort from the JHS; 4) the South German Myocardial Infarction study (dbGaP study accession phs000916.v1.p1); 5) the Ottawa Heart Study (OHS) (dbGaP study accession phs000806.v1.p1); 6) the Precocious Coronary Artery Disease (PROCARDIS) study (dbGaP Study Accession phs000883.v1.p1) ; 7) the Pakistan Risk of Myocardial Infarction Study (PROMIS) (dbGaP study accession phs000917.v1.p1); 8) the Registre Gironí del COR (Gerona Heart Registry or REGICOR) study (dbGaP study accession phs000902.v1.p1); 9) the Leicester Myocardial Infarction study (dbGaP study accession phs001000.v1.p1); 10) the BioImage study (dbGaP study accession phs001058.v1.p1); and 11) the North German Myocardial Infarction study (dbGaP Study Accession phs000990.v1.p1).
Predicted damaging ACVR1C variants in the exome sequencing analysis were defined as those that resulted in loss of function of the protein (nonsense mutations that resulted in early termination of ACVR1C, frameshift mutations due to insertions or deletions of DNA, or splice-site mutations that result in an incorrectly spliced protein) or those labeled as damaging by each of five different algorithms (LRT score, MutationTaster, PolyPhen-2 HumDiv, PolyPhen-2 HumVar, and SIFT), as previously described (19,26). The Variant Effect Predictor algorithm was used to annotate predicted damaging variants (27).
Statistical Analysis
In UK Biobank, WHRadjBMI was derived through inverse normal transformation of waist-to-hip ratio after adjustment for age, sex, and BMI (as in the GIANT Consortium [16]). In UK Biobank, linear regression was used to estimate the association of variants with WHRadjBMI. All UK Biobank analyses included adjustment for age, sex, 10 principal components of ancestry, and a dummy variable for the array type used in genotyping. Logistic regression was used to estimate the association of variants with type 2 diabetes. Estimates of the association of each variant with type 2 diabetes in UK Biobank were pooled with estimates from the ExTexT2D Consortium using inverse variant–weighted fixed-effects meta-analysis.
In the primary analysis of 405,569 individuals for WHRadjBMI, we had 80% power to detect a minimum effect of 0.05 SD with a minor allele frequency of 5% at genome-wide significance (P < 5 × 10−8). In the primary analysis of 95,978 case subjects with type 2 diabetes and 646,985 control subjects, we had 80% power to detect a minimum odds ratio (OR) of 1.05 at P < 5 × 10−8. With a minor allele frequency of 1%, we had 80% power to detect a minimum effect of 0.1 SD for WHRadjBMI and an OR of 1.10 for type 2 diabetes at P < 5 × 10−8.
To estimate the overall association of variation in ACVR1C with WHRadjBMI, we pooled across all variants using a gene risk score, weighted by the square root of allele frequency (estimating a weighted mean effect of ACVR1C variants on WHRadjBMI) (28). To estimate the overall association of variation in ACVR1C with type 2 diabetes, we pooled across all variants in a gene risk score, weighted by the association of each variant with WHRadjBMI (29).
For analysis of exome sequencing data, logistic regression was performed with adjustment for sex, five principal components of ancestry, and a dummy variable for each cohort to estimate the association of predicted damaging variants with type 2 diabetes. Estimates from MIGen were pooled with estimates from the T2D-GENES Consortium using inverse variance–weighted fixed-effects meta-analysis.
For the phenome-wide association study, all four ACVR1C variants were pooled in a gene risk score in UK Biobank, as previously described (25,30). For each individual in UK Biobank, the ACVR1C variants associated with lower WHRadjBMI were weighted by their effect on WHRadjBMI and summed. The association of this gene risk score with 31 different diseases in UK Biobank and three metabolic traits was tested using logistic regression with adjustment for age, sex, 10 principal components of ancestry, and a dummy variable for the array type used in genotyping. Although exploratory due to the low number of case subjects for certain diseases (e.g., 1,707 case subjects for cervical cancer), we conducted this analysis to detect possible adverse associations of ACVR1C with diseases that could allow for prediction of adverse effects of pharmacologic inhibition of ACVR1C.
Analyses were performed using R version 3.2.3 (R Project for Statistical Computing).
Results
Exome-Wide Association Study of Body Fat Distribution in UK Biobank
Among 405,569 participants in UK Biobank, 54% were female, the median age was 57 years, and the median waist-to-hip ratio, measured at enrollment, was 0.87 (Table 1). One SD in waist-to-hip ratio corresponded to an absolute change of 0.09. In an analysis of 614,012 coding variants in UK Biobank, no evidence of genomic inflation was observed (λ 1.08) (Supplementary Fig. 1).
Table 1.
Baseline characteristics of participants in UK Biobank
| All participants (N = 405,569) | Type 2 diabetes case subjects (N = 20,458) | Control subjects without diabetes (N = 385,111) | |
|---|---|---|---|
| Age, years | 57 ± 8.1 | 61 ± 6.9 | 57 ± 8.1 |
| Female | 218,376 (54) | 7,801 (38) | 210,575 (55) |
| UK BiLEVE array | 48,625 (12.0) | 3,097 (15) | 45,528 (12) |
| BMI, kg/m2 | 27 ± 4.8 | 32 ± 5.9 | 27 ± 4.6 |
| Waist-to-hip ratio | 0.87 ± 0.09 | 0.95 ± 0.08 | 0.87 ± 0.09 |
Data are mean ± SD or n (%).
We identified 16 low-frequency variants (<5%) associated with WHRadjBMI outside of known loci (Supplementary Table 2) (16). We identified an additional 43 novel common variants (frequency >5%) associated with WHRadjBMI (Supplementary Table 3). We identified 94 independent variants in total, including variants at known loci (Supplementary Data). Of 59 novel coding variants, 34 variants were available in the GIANT Consortium for replication (16). A strong correlation in the effect sizes of the association of variants with WHRadjBMI in UK Biobank and GIANT was observed (R2 = 0.87). A total of 24 variants were independently associated with WHRadjBMI in GIANT (P < 0.05) (Supplementary Table 4). Of the 10 variants that were not significantly associated with WHRadjBMI in GIANT (P > 0.05), only 3 variants exhibited evidence of heterogeneity between estimates in UK Biobank and GIANT, with the remaining 7 variants showing similar estimates of association in UK Biobank and GIANT (although not reaching significance in GIANT) (Supplementary Table 4).
The lead novel low-frequency variant was a missense variant in PNPLA2 (rs140201358, Asn252Lys) that associated with elevated WHRadjBMI (0.09 SD, P = 3.2 × 10−19). PNPLA2 encodes adipocyte triglyceride lipase, which hydrolyzes triglycerides in adipose tissue to mobilize fat stores (31). Two low-frequency missense variants in ABHD15, which encodes alpha/beta hydrolase domain-containing protein 15, were found to be associated with elevated WHRadjBMI. ABHD15 is also highly expressed in adipocytes and has been reported to mediate insulin-induced suppression of lipolysis in adipocytes (32). We also identified a low-frequency variant in a known locus (CALCRL Leu87Pro, 0.1% frequency) associated with lower WHRadjBMI (−0.14 SD, P = 1.9 × 10−11). A common noncoding variant in the CALCRL locus, encoding calcitonin receptor-like receptor, was previously identified in the GIANT Consortium as associated with WHRadjBMI (16). The identification of an independent low-frequency missense variant in the gene suggests that CALCRL may be the causal gene at this locus.
Variation Leading to Lower WHRadjBMI: The ACVR1C Locus
We next focused on novel variant alleles leading to lower WHRadjBMI. The lead novel low-frequency variant associated with lower WHRadjBMI lay within the gene ACVR1C. ACVR1C Asn150His (allele frequency 1.1%) associated with 0.09 SD lower WHRadjBMI (P = 3.4 × 10−17). An independent missense variant in ACVR1C, Ile195Thr (AF 0.4%), also associated with lower WHRadjBMI (0.15 SD, P = 1.0 × 10−9).
Upon conditioning on these two variants, we identified an additional coding variant: Ile482Val (AF 7%), which associated with 0.019 SD lower WHRadjBMI (P = 1.6 × 10−5), and rs72927479, a noncoding variant, for which the minor G allele (AF 5%) was associated with 0.035 SD lower WHRadjBMI (P = 2.6 × 10−12) (Table 2). Despite being an independent signal for WHRadjBMI, the noncoding variant rs72927479 is nominally correlated with Ile482Val (r2 = 0.06 in UK Biobank). No other variants were correlated with one another in UK Biobank (all r2 < 0.001).
Table 2.
Association of variants in ACVR1C with WHRadjBMI and with type 2 diabetes
| Variant | Minor allele frequency (%) | WHRadjBMI |
Type 2 diabetes |
||
|---|---|---|---|---|---|
| β (95% CI) | P value | OR (95% CI) | P value | ||
| Asn150His |
1.1 |
−0.089 (−0.11, −0.067) |
3.4 × 10−17 |
0.88 (0.83, 0.94) |
8.7 × 10−5 |
| Ile195Thr |
0.2 |
−0.15 (−0.09, 0.19) |
1.0 × 10−9 |
0.79 (0.67, 0.93) |
0.005 |
| Ile482Val |
7.2 |
−0.019 (−0.01, −0.027) |
1.6 × 10−5 |
0.95 (0.93, 0.97) |
4.8 × 10−6 |
| rs72927479 | 5.1 | −0.035 (−0.045, −0.025) | 2.6 × 10−12 | 0.93 (0.89, 0.97) | 6.0 × 10−4 |
Estimates for WHRadjBMI were derived through linear regression analysis in UK Biobank. Estimates for type 2 diabetes were derived through meta-analysis of UK Biobank and the DIAGRAM ExTexT2D Consortium.
Pooling across all four variants with weighting by square root of allele frequency, ACVR1C variation was associated with lower WHRadjBMI in UK Biobank (−0.07 SD, P = 2.6 × 10−35). To replicate this finding, we pooled data from the GIANT Consortium (in which the Ile482Val variant was available, n = 224,156) with data from ARIC and Framingham Heart Study data sets (in which Asn150His, Ile195Thr, and rs72927479 variants were available, n = 13,704). Pooling across all four variants in these three replication studies, variation in ACVR1C was associated with reduced WHRadjBMI (−0.07 SD, P = 0.0005) (Supplementary Table 5).
Examining other anthropometric traits in UK Biobank, variation in ACVR1C was associated with elevated hip circumference (0.035 SD, P = 3 × 10−8) and nominally elevated BMI (0.02 SD, P = 0.002) and was unassociated with waist circumference (−0.007 SD, P = 0.20) or height (0.005 SD, P = 0.23). In the Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) analysis of insulin resistance (HOMA-IR), Ile482Val (the only ACVR1C variant available for analysis) was unassociated with HOMA-IR (−1.1%, P = 0.17) (33).
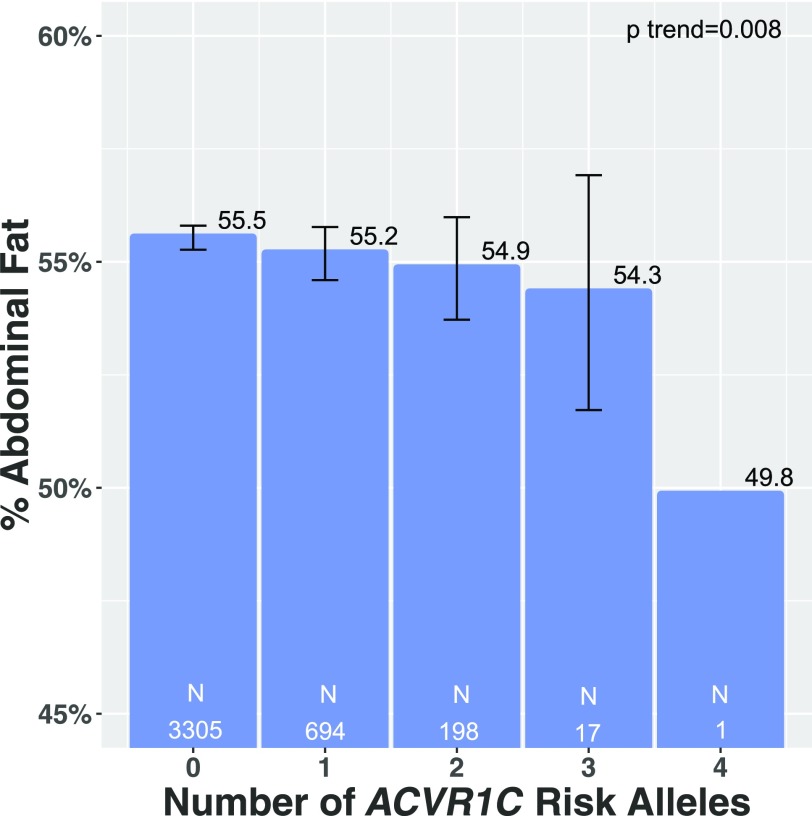
We examined whether ACVR1C variation also associated with direct imaging measurement of abdominal obesity. A total of 4,215 participants in UK Biobank underwent DEXA imaging to estimate abdominal fat mass. Carriers of ACVR1C variants had lower percent abdominal fat (P = 0.008) (Fig. 1). When one outlying individual with four ACVR1C variants was excluded (Fig. 1), carriers of ACVR1C continued to have significantly lower percent abdominal fat (P = 0.009).
Figure 1.
Across four ACVR1C genetic variants, association of number of WHRadjBMI-lowering alleles with mean directly measured abdominal fat (percent abdominal fat of total body fat) in 4,215 participants who underwent DEXA scan in UK Biobank, adjusted for age, sex, 10 principal components of ancestry, and array type. Number of participants in each group is displayed in white for each bar.
Association of Variants in ACVR1C With Type 2 Diabetes
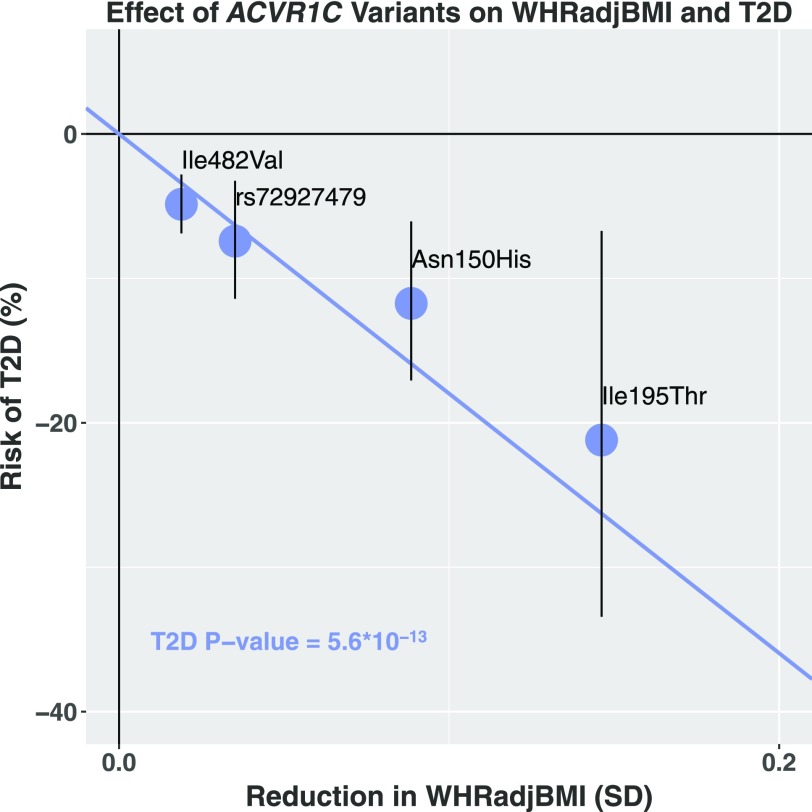
Genetic predisposition to increased WHRadjBMI strongly predisposes to type 2 diabetes (12). We therefore examined whether variants in ACVR1C that lower WHRadjBMI adiposity protect against type 2 diabetes. In a combined analysis of UK Biobank and the DIAGRAM Consortium, all four ACVR1C variants were found to independently protect against type 2 diabetes (OR 0.88, P = 8.7 × 10−5 for Asn150His; OR 0.79, P = 0.005 for Ile195Thr; OR 0.95, P = 4.8 × 10−6 for Ile482Val; OR 0.93, P = 0.0006 for rs72927479) (Table 1). Pooling across all four variants, a 0.2 SD decrease in WHRadjBMI through ACVR1C was associated with a 30% lower risk of type 2 diabetes (OR 0.70, 95% CI 0.63, 0.77; P = 5.6 × 10−13) (Fig. 2). When we excluded the noncoding variant rs72927479, which is nominally correlated with Ile482Val (r2 = 0.06 in UK Biobank), the ACVR1C gene risk score remained associated with risk of type 2 diabetes (OR 0.71, 95% CI 0.64, 0.79; P = 1.8 × 10−10)
Figure 2.
Association of four variants in ACVR1C with WHRadjBMI (x-axis) and type 2 diabetes (T2D) (y-axis).
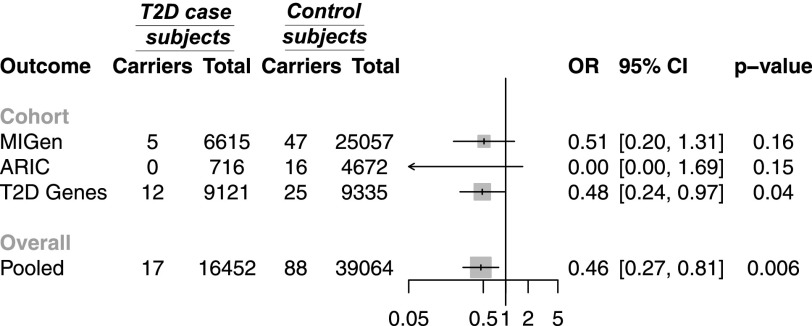
In three independent data sets with exome sequence data—MIGen, ARIC, and the T2D-GENES Consortium—we examined whether variants predicted to damage ACVR1C gene function protect against type 2 diabetes. The nine exons of the ACVR1C gene were sequenced in 55,516 individuals. A total of 105 predicted damaging variants were identified (Supplementary Tables 6–8). Among 16,452 case subjects with type 2 diabetes, the frequency of predicted damaging variants in ACVR1C was 0.1% (17) compared with 0.2% (88) among 39,064 control subjects without diabetes. Overall, carrying a predicted damaging variant in ACVR1C was associated with 54% lower risk of type 2 diabetes (OR 0.46, 95% CI 0.27, 0.81; P = 0.006) (Fig. 3). When we excluded 47 carriers of I195T (annotated as a predicted damaging variant by five of five algorithms) in the exome sequencing analysis, carriers of predicted damaging variants in ACVR1C remained protected from type 2 diabetes (OR 0.48, 95% CI 0.24, 0.97; P = 0.04).
Figure 3.
Association of predicted damaging variants in ACVR1C with type 2 diabetes (T2D) from sequences in MIGen, ARIC, and the T2D-GENES Consortium (T2D Genes).
To further examine whether loss of ACVR1C function lowers WHRadjBMI, we examined whether the noncoding variant rs72927479 associates with ACVR1C expression. The minor allele of rs72927479 (G, frequency 5%) associated with lower WHRadjBMI (β −0.035, P = 2.6 × 10−12) and type 2 diabetes (OR 0.93, P = 0.0006). In the Genotype-Tissue Expression (GTEx) data set (34), the minor allele of rs72927479 associated with reduced expression of ACVR1C in subcutaneous adipose tissue (P = 0.02) and pancreas (P = 0.02) (Supplementary Fig. 2).
Phenome-Wide Association Study of ACVR1C in UK Biobank
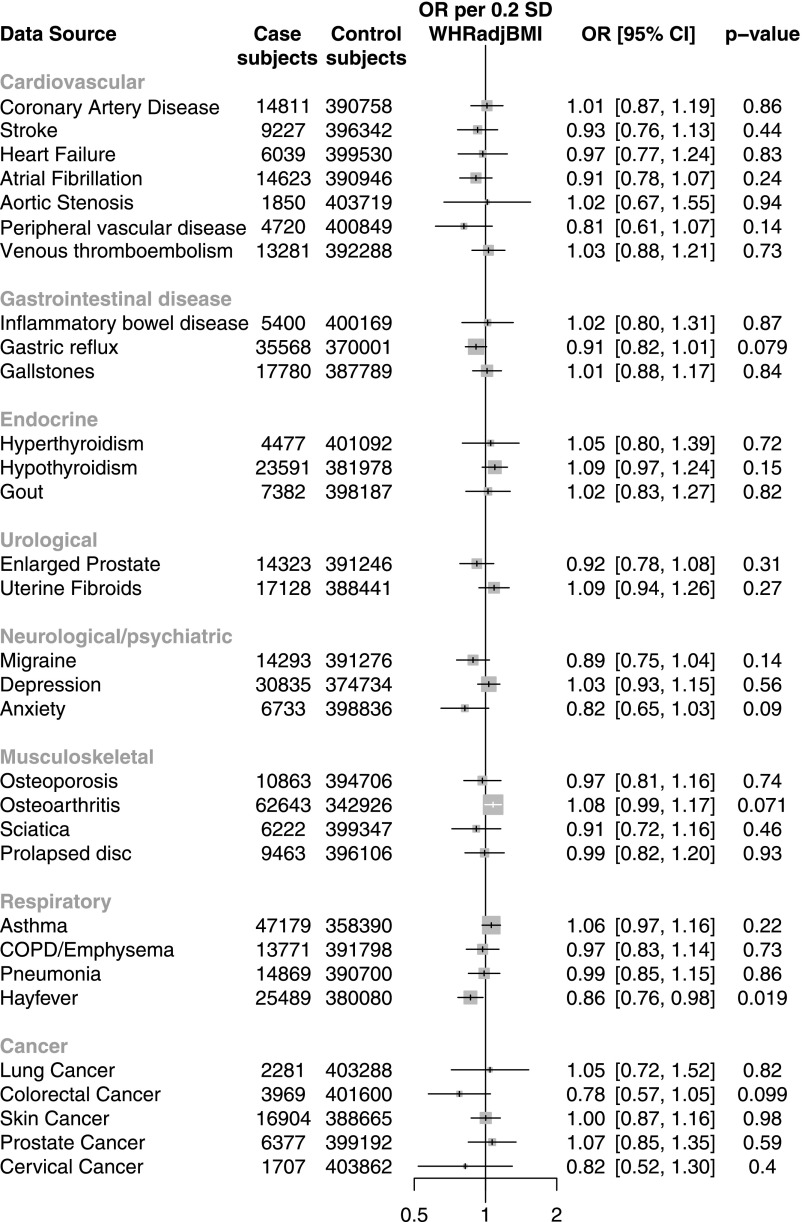
To anticipate whether ACVR1C inhibition may be associated with on-target adverse effects, we conducted a phenome-wide association study of 31 disease phenotypes in UK Biobank. We did not observe any significant associations between the ACVR1C gene risk score and the 31 diseases analyzed (Fig. 4). The ACVR1C gene risk score was unassociated with coronary artery disease (OR 1.01, P = 0.86). We also examined whether ACVR1C variation associates with three metabolic traits currently available in UK Biobank: urinary albumin, systolic blood pressure, and diastolic blood pressure. While the ACVR1C gene risk score did not significantly associate with urinary albumin levels, it associated with significantly lower diastolic blood pressure (−0.6 mmHg, P = 0.0004) and nominally lower systolic blood pressure (−0.6 mmHg, P = 0.03).
Figure 4.
Association of ACVR1C gene risk score with 31 disease phenotypes in UK Biobank. COPD, chronic obstructive pulmonary disease.
Discussion
In this study, four genetic variants in ACVR1C, ranging in frequency from 0.2 to 7.2%, independently associated with lower WHRadjBMI and protected against type 2 diabetes. Furthermore, damaging variants in ACVR1C protected against type 2 diabetes in an analysis of exome sequences from 55,516 individuals. An ACVR1C gene risk score did not associate with any of 31 additional diseases in UK Biobank but did nominally associate with lower blood pressure.
These results permit several conclusions. First, pharmaceutical inhibition of ACVR1C may be useful in the treatment of type 2 diabetes. ACVR1C encodes ALK7, a transforming growth factor-β family receptor highly expressed on pancreatic islet cells (35,36) and adipocytes (37). Overexpression of AKL7 induces growth inhibition and apoptosis of pancreatic β-cells (36,38), suggesting that it is a negative regulator of β-cell mass. A number of findings from model systems also suggest that ALK7 may be useful as a therapeutic target for abdominal obesity, type 2 diabetes, and other metabolic diseases. ACVR1C-deficient mice have been reported to have reduced body fat when fed a high-fat diet (37,39) and have improved glucose tolerance and insulin sensitivity when obese (37). Chemical inhibition of ACVR1C has also been shown to reduce fat accumulation and increase lipolysis in mice (40). Rats with streptozocin-induced diabetes show elevated ALK7 expression, and shRNA knockdown of ACVR1C reduces arterial stiffness in this model (41). In combination with the human genetic results presented here, these findings suggest that inhibition of ACVR1C may prove useful to modify body fat distribution and lower risk for type 2 diabetes.
Second, the lack of association of the ACVR1C gene risk score with 31 different diseases in UK Biobank suggests that therapeutic ACVR1C inhibition may not have adverse on-target effects. In the Exome Aggregation Consortium (ExAC), ACVR1C is tolerant of loss-of-function variants, with 18 of 66,720 individuals of European ancestry carrying an early stop codon (Leu32Ter) in ACVR1C (42). In combination with the phenome-wide association study presented here, these findings suggest that ACVR1C could be safely inhibited. However, due to the small number of case subjects for many of the analyzed diseases and the multiple diseases tested in the phenome-wide association study, modest associations of ACVR1C variation with the analyzed phenotypes cannot excluded. Furthermore, many common adverse effects of therapeutics, such as elevations in liver function enzymes, could not be analyzed due to a lack of available data in UK Biobank. In particular, ALK7-deficient mice have been reported to have lengthened QT intervals (43), a phenotype that was unavailable for analysis in UK Biobank. The association of the ACVR1C gene risk score with nominally lower diastolic and systolic blood pressure suggests that ACVR1C inhibition may have the additional benefit of lowering blood pressure. However, this finding, which did not reach genome-wide significance, requires replication in independent data sets.
The primary strength of the analysis is the use of multiple data sources to replicate the association of ACVR1C variation with WHRadjBMI and type 2 diabetes. We demonstrated associations of ACVR1C with WHRadjBMI and type 2 diabetes to greater than genome-wide significance (P = 2.6 × 10−35 for WHRadjBMI and P = 5.6 × 10−13 for type 2 diabetes). We further showed that variants leading to loss of ACVR1C function protected against diabetes in an analysis of predicted damaging variants in exome sequences of 55,516 individuals. A primary limitation of the analysis is that we did not experimentally characterize the analyzed variants. The consistent protective associations observed with three different ACVR1C missense variants, with a noncoding variant that reduces ACVR1C expression, and with variants predicted to truncate the ACVR1C protein or predicted to damage ACVR1C function by five different algorithms suggest that variants leading to loss of ACVR1C function protect against type 2 diabetes. However, experimental demonstration that ACVR1C variants that lower WHRadjBMI and protect against type 2 diabetes actually reduce ALK7 receptor function is necessary before it can be concluded that ACVR1C deficiency will protect against type 2 diabetes. A second limitation is that pharmaceutical ACVR1C inhibition may be associated with off-target effects that cannot be characterized in a human genetic study.
In summary, variants predicted to damage ACVR1C gene function lower WHRadjBMI and protect against type 2 diabetes. These findings provide human genetic validation for the ACVR1C gene as a therapeutic target for type 2 diabetes.
Supplementary Material
Article Information
Funding and Duality of Interest. This research has been conducted using the UK Biobank resource, application 7089. This work was funded by the National Institutes of Health (R01 HL127564 to S.K.), which had no involvement in the design and conduct of the study; the collection, analysis, and interpretation of the data; or the preparation, review, and approval of the manuscript. This project was also conducted using the Type 2 Diabetes Knowledge Portal resource, which is funded by the Accelerating Medicines Partnership. The REGICOR study was supported by the Spanish Ministry of Economy and Innovation through the Carlos III Health Institute (Red Investigación Cardiovascular RD12/0042, PI09/90506), European Regional Development Fund (ERDF), and the Catalan Research and Technology Innovation Interdepartmental Commission (2014SGR240). Samples for the Leicester cohort were collected as part of projects funded by the British Heart Foundation (British Heart Foundation Family Heart Study, British Heart Foundation grant RG2000010, United Kingdom Aneurysm Growth Study [UKAGS], British Heart Foundation grant CS/14/2/30841) and the National Institute for Health Research (NIHR) (NIHR Leicester Cardiovascular Biomedical Research Unit Biomedical Research Informatics Centre for Cardiovascular Science, IS_BRU_0211_20033). N.J.S. is supported by the British Heart Foundation and is an NIHR Senior Investigator. The Northern German Myocardial Infarction Study is supported by the German Federal Ministry of Education and Research (BMBF) in the context of the e:Med program (e:AtheroSysMed) and the FP7 European Union project CVgenes@target (261123). Additional grants were received from the Fondation Leducq (CADgenomics: Understanding Coronary Artery Disease Genes, 12CVD02). This study was also supported through the Deutsche Forschungsgemeinschaft cluster of excellence “Inflammation at Interfaces” and SFB 1123. The Italian ATVB study was supported by a grant from RFPS-2007-3-644382 and Programma di ricerca Regione-Università 2010-2012 Area 1–Strategic Programmes–Regione Emilia-Romagna. Funding for ESP was provided by RC2 HL103010 (HeartGO), RC2 HL102923 (LungGO), and RC2 HL102924 (WHISP). Exome sequencing was performed through RC2 HL102925 (BroadGO) and RC2 HL102926 (SeattleGO). The JHS is supported by contracts HHSN268201300046C, HHSN268201300047C, HHSN268201300048C, HHSN268201300049C, and HHSN268201300050C from the National Heart, Lung, and Blood Institute and the National Institute on Minority Health and Health Disparities. J.G.W. is supported by U54GM115428 from the National Institute of General Medical Sciences. Exome sequencing in ATVB, PROCARDIS, Ottawa, PROMIS, South German Myocardial Infarction Study, and the JHS was supported by 5U54HG003067 (to S.G.). A.V.K. is supported by a K08 from the National Human Genome Research Institute (K08HG010155) and a Junior Faculty Award from the National Lipid Association and has received consulting fees from Amarin. P.N. reports funding from the John S. LaDue Memorial Fellowship at Harvard Medical School and has received consulting fees from Amarin. S.K. is supported by a research scholar award from Massachusetts General Hospital, the Donovan Family Foundation, and R01 HL127564; has received a research grant from Bayer Healthcare and consulting fees from Merck, Novartis, Sanofi, AstraZeneca, Alnylam Pharmaceuticals, Leerink Partners, Noble Insights, MedGenome, Aegerion Pharmaceuticals, Regeneron Pharmaceuticals, Quest Diagnostics, Color Genomics, Genomics PLC, and Eli Lilly and Company; and holds equity in San Therapeutics, Catabasis Pharmaceuticals, and Endcadia. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. C.A.E. and S.K. contributed to study concept and design. C.A.E., A.V.K., M.C., D.S., S.G., and S.K. contributed to acquisition, analysis, or interpretation of data. C.A.E., A.V.K., and S.K. drafted the manuscript. S.K. provided administrative, technical, and material support. All authors critically revised the manuscript for important intellectual content. C.A.E. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db18-0857/-/DC1.
References
- 1.Edwards SL, Beesley J, French JD, Dunning AM. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet 2013;93:779–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abifadel M, Varret M, Rabès J-P, et al. . Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet 2003;34:154–156 [DOI] [PubMed] [Google Scholar]
- 3.Cohen JC, Boerwinkle E, Mosley TH, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med 2006;354:1264–1272 [DOI] [PubMed]
- 4.Sabatine MS, Giugliano RP, Keech AC, et al.; FOURIER Steering Committee and Investigators. Evolocumab and clinical outcomes in patients with cardiovascular disease. N Engl J Med 2017;376:1713–1722 [DOI] [PubMed]
- 5.Stitziel NO, Khera AV, Wang X, et al.; PROMIS and Myocardial Infarction Genetics Consortium Investigators . ANGPTL3 deficiency and protection against coronary artery disease. J Am Coll Cardiol 2017;69:2054–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dewey FE, Gusarova V, Dunbar RL, et al. Genetic and pharmacologic inactivation of ANGPTL3 and cardiovascular disease. N Engl J Med 2017;377:211–221 [DOI] [PMC free article] [PubMed]
- 7.Graham MJ, Lee RG, Brandt TA, et al. Cardiovascular and metabolic effects of ANGPTL3 antisense oligonucleotides. N Engl J Med 2017;377:222–232 [DOI] [PubMed]
- 8.Pollin TI, Damcott CM, Shen H, et al. . A null mutation in human APOC3 confers a favorable plasma lipid profile and apparent cardioprotection. Science 2008;322:1702–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crosby J, Peloso GM, Auer PL, et al.; TG and HDL Working Group of the Exome Sequencing Project, National Heart, Lung, and Blood Institute . Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N Engl J Med 2014;371:22–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jørgensen AB, Frikke-Schmidt R, Nordestgaard BG, Tybjærg-Hansen A. Loss-of-function mutations in APOC3 and risk of ischemic vascular disease. N Engl J Med 2014;371:32–41 [DOI] [PubMed]
- 11.Gaudet D, Alexander VJ, Baker BF, et al. Antisense inhibition of apolipoprotein C-III in patients with hypertriglyceridemia. N Engl J Med 2015;373438–447 [DOI] [PubMed]
- 12.Emdin C, Khera AV, Natarajan P, et al. Genetic association of waist-to-hip ratio with cardiometabolic traits, type 2 diabetes, and coronary heart disease. JAMA 2017;317:626–634 [DOI] [PMC free article] [PubMed]
- 13.Yusuf S, Hawken S, Ounpuu S, et al.; INTERHEART Sudy Investigators. Obesity and the risk of myocardial infarction in 27,000 participants from 52 countries: a case-control study. Lancet 2005;366:1640–1649 [DOI] [PubMed]
- 14.Dale CE, Fatemifar G, Palmer TM, et al.; UCLEB Consortium; METASTROKE Consortium . Causal associations of adiposity and body fat distribution with coronary heart disease, stroke subtypes, and type 2 diabetes mellitus: a Mendelian randomization analysis. Circulation 2017;135:2373–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shungin D, Winkler TW, Croteau-Chonka DC, et al.; ADIPOGen Consortium; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GEFOS Consortium; GENIE Consortium; GLGC; ICBP; International Endogene Consortium; LifeLines Cohort Study; MAGIC Investigators; MuTHER Consortium; PAGE Consortium; ReproGen Consortium . New genetic loci link adipose and insulin biology to body fat distribution. Nature 2015;518:187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeBoever C, Tanigawa Y, Lindholm ME, et al. . Medical relevance of protein-truncating variants across 337,205 individuals in the UK Biobank study. Nat Commun 2018;9:1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahajan A, Wessel J, Willems SM, et al.; ExomeBP Consortium; MAGIC Consortium; GIANT Consortium . Refining the accuracy of validated target identification through coding variant fine-mapping in type 2 diabetes. Nat Genet 2018;50:559–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Do R, Stitziel NO, Won H-H, et al.; NHLBI Exome Sequencing Project . Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature 2015;518:102–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khera AV, Won H-H, Peloso GM, et al. . Diagnostic yield and clinical utility of sequencing familial hypercholesterolemia genes in patients with severe hypercholesterolemia. J Am Coll Cardiol 2016;67:2578–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuchsberger C, Flannick J, Teslovich TM, et al. . The genetic architecture of type 2 diabetes. Nature 2016;536:41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emdin CA, Khera AV, Klarin D, et al.; American Heart Association . Phenotypic consequences of a genetic predisposition to enhanced nitric oxide signaling. Circulation 2018;137:222–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.ARIC Investigators. The Atherosclerosis Risk in Communities (ARIC) study: design and objectives. Am J Epidemiol 1989;129:687–702 [PubMed] [Google Scholar]
- 24.Khera AV, Chaffin M, Aragam K, et al. Genome-wide polygenic score to identify a monogenic risk-equivalent for coronary disease. Nat Genet 2018;50:1219–1224 [DOI] [PMC free article] [PubMed]
- 25.Emdin CA, Khera AV, Natarajan P, et al.; CHARGE–Heart Failure Consortium; CARDIoGRAM Exome Consortium . Phenotypic characterization of genetically lowered human lipoprotein(a) levels. J Am Coll Cardiol 2016;68:2761–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khera AV, Won H-H, Peloso GM, et al.; Myocardial Infarction Consortium; DiscovEHR Study Group, CARDIoGRAM Exome Consortium; Global Lipids Genetics Consortium. Association of rare and common variation in the lipoprotein lipase gene with coronary artery disease. JAMA 2017;317:937–946 [DOI] [PMC free article] [PubMed]
- 27.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 2010;26:2069–2070 [DOI] [PMC free article] [PubMed]
- 28.Park J-H, Gail MH, Weinberg CR, et al. . Distribution of allele frequencies and effect sizes and their interrelationships for common genetic susceptibility variants. Proc Natl Acad Sci U S A 2011;108:18026–18031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu DJ, Peloso GM, Yu H, et al.; Charge Diabetes Working Group; EPIC-InterAct Consortium; EPIC-CVD Consortium; GOLD Consortium; VA Million Veteran Program . Exome-wide association study of plasma lipids in >300,000 individuals. Nat Genet 2017;49:1758–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emdin CA, Klarin D, Natarajan P, Florez JC, Kathiresan S, Khera AV; CARDIOGRAM Exome Consortium . Genetic variation at the sulfonylurea receptor, type 2 diabetes, and coronary heart disease. Diabetes 2017;66:2310–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zimmermann R, Strauss JG, Haemmerle G, et al. . Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science 2004;306:1383–1386 [DOI] [PubMed] [Google Scholar]
- 32.Xia W, Pessentheiner AR, Hofer DC, et al. . Loss of ABHD15 impairs the anti-lipolytic action of insulin by altering PDE3B stability and contributes to insulin resistance. Cell Reports 2018;23:1948–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dupuis J, Langenberg C, Prokopenko I, et al.; DIAGRAM Consortium; GIANT Consortium; Global BPgen Consortium; Anders Hamsten on behalf of Procardis Consortium; MAGIC investigators . New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 2010;42:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015;348:648–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bondestam J, Huotari MA, Morén A, et al. cDNA cloning, expression studies and chromosome mapping of human type I serine/threonine kinase receptor ALK7 (ACVR1C). Cytogenet Cell Genet 2001;95:157–162 [DOI] [PubMed]
- 36.Zhang N, Kumar M, Xu G, et al. Activin receptor-like kinase 7 induces apoptosis of pancreatic beta cells and beta cell lines. Diabetologia 2006;49:506–518 [DOI] [PubMed]
- 37.Yogosawa S, Mizutani S, Ogawa Y, Izumi T. Activin receptor-like kinase 7 suppresses lipolysis to accumulate fat in obesity through downregulation of peroxisome proliferator-activated receptor γ and C/EBPα. Diabetes 2013;62:115–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao F, Huang F, Tang M, et al. . Nodal induces apoptosis through activation of the ALK7 signaling pathway in pancreatic INS-1 β-cells. Am J Physiol Endocrinol Metab 2012;303:E132–E143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andersson O, Korach-Andre M, Reissmann E, Ibáñez CF, Bertolino P. Growth/differentiation factor 3 signals through ALK7 and regulates accumulation of adipose tissue and diet-induced obesity. Proc Natl Acad Sci U S A 2008;105:7252–7256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo T, Marmol P, Moliner A, et al. . Adipocyte ALK7 links nutrient overload to catecholamine resistance in obesity. eLife 2014;3:e03245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li W-B, Zhao J, Liu L, et al. . Silencing of activin receptor-like kinase 7 alleviates aortic stiffness in type 2 diabetic rats. Acta Diabetol 2015;52:717–726 [DOI] [PubMed] [Google Scholar]
- 42.Lek M, Karczewski KJ, Minikel EV, et al.; Exome Aggregation Consortium . Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ying S, Cao H, Hu H, Wang X, Tang Y, Huang C. Alk7 depleted mice exhibit prolonged cardiac repolarization and are predisposed to ventricular arrhythmia. PLoS One 2016;11:e0149205 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.