Abstract
Background: Conventional antibiotic agents are overused, leading to decreased efficacy because of a rising incidence in antimicrobial resistance. Further, conventional antibiotic agents result in widespread effects to human microbiota, which can lead directly to adverse events such as Clostridium difficile infection.
Methods: This review provides a narrative summary of anti-sense therapies, an approach to managing bacterial infections by pursuing specific molecular targets that disrupt the flow of information from deoxyribonucleic acid to ribonucleic acid to protein, leading to the loss of bacterial functions. Included in this article is the rationale for this approach, the current data supporting its further investigation, and the challenges and future directions in this area of research.
Results: There is a compelling proof-of-concept against both gram-positive and gram-negative organisms to commend the use of modified anti-sense oligonucleotides as antimicrobial therapy. There are data demonstrating that anti-sense therapies are capable of killing bacteria, silencing antimicrobial resistance mechanisms to restore sensitivity to conventional antibiotic agents, and to target virulence pathways such as biofilm production. Further, these drugs have a significantly greater degree of organismal specificity, limiting antibiotic-associated diarrhea and lowering the risk of antibiotic-related infections such as C. difficile infection.
Conclusions: Anti-sense therapies show promise as a new class of antibiotic agents, providing molecular precision that leads to specific targeting of bacterial species and bacterial functions, including virulence mechanisms beyond the reach of current antibiotic agents. Further, changing the sequence of an anti-sense oligonucleotide provides a method of dealing with antimicrobial resistance that is more time- and cost-flexible than the available options with current conventional antibiotic agents.
Keywords: antibiotics, anti-sense, Clostridium difficile
Conventional antibiotic agents are a near-ubiquitous element of both inpatient and outpatient medicine, accounting for $10 billion in annual healthcare expenditures in 2009 in the United States [1]. When considering the unnecessary use of these drugs combined with the inappropriate selection, dosage, or duration of an antibiotic agent, total inappropriate antibiotic use approaches 50% of all antibiotic courses prescribed in the United States [1]. It is this non-evidence based utilization of antibiotic agents that has promoted the world-wide crisis of antimicrobial resistance (AMR), defined [2] by the World Health Organization (WHO) as “the ability of a micro-organism (like bacteria, viruses, and some parasites) to stop an antimicrobial (such as antibiotics, antivirals and antimalarials) from working against it.” It is estimated that AMR contributes an annual $2.2 billion in healthcare expenditures [3] and, more importantly, it represents a direct threat to life for patients, even those with common bacterial infections.
Because of its prevalence and severity, and with no immediate solution, the WHO has declared [2] AMR to be one of the top three most important public health issues. Simultaneously, there has been a lack of new antibiotic development [4] during the previous two decades to keep pace with AMR, resulting in the increased use of currently available antibiotic agents with diminishing returns secondary to selective pressures that only increase the proportion of resistant organisms.
The development of AMR is a predictable result of bacterial exposure to antibiotic agents, because most of these drugs are structurally analogous to naturally occurring molecules [5], allowing bacteria to evolve mechanisms to mitigate the activity of antibiotic agents via exposure to their naturally occurring counterparts. The two most important bacterial resistance mechanisms involve mutational resistance [6] and horizontal gene transfer [7]. The former represents an acquired genetic variant arising from replication errors, one that can provide a selective advantage to bacteria being exposed to an antibiotic agent. The most common acquired beneficial functions introduced to bacteria by mutational resistance include the ability to modify the target of the antibiotic agent, to alter or destroy the antibiotic agent, or to either decrease the uptake of or actively remove the drug. In the case of horizontal gene transfer, the role of bacteriophages has been instrumental in driving bacterial evolution through transduction, although conjugative mobile genetic elements such as transposons as well as the direct uptake of genetic material from the environment (transformation) also contribute to the acquisition of new functions.
When considering the sheer number of phages in the human gut and the environment, coupled with the rapidity of bacterial replication, AMR is expectedly both frequent and directly proportional to bacterial exposure to antibiotic agents. Depending on the exact type of AMR that emerges, minor structural modifications to an antibiotic agent may not be adequate to address resistance, potentially disqualifying a drug or even an entire class of drugs from further use. This common scenario requires the creation of an entirely new drug, representing an expensive and lengthy drug development process.
Conventional antibiotic agents have two other limitations best demonstrated by using Clostridium difficile infection (CDI), the most common [8] nosocomial bacterial infection in the United States, as a paradigm. First, conventional antibiotic agents lack organismal specificity, leading to widespread ecologic changes involving gut microbiota that not only produce antibiotic-associated diarrhea, but also in the case of CDI, actually promote [9] the development of this infection because of the loss of beneficial biomass from the gut. Second, virulence mechanisms such as sporulation, biofilm production, and envenomation are promising therapeutic aims that are not directly targetable by conventional antibiotic agents; in fact, conventional antibiotic agents often serve as environmental cues [10–12] for the enrichment of each of these virulence pathways.
Antimicrobial resistance MR is a global problem capable of transforming even mundane infections into life-threating diseases. Conventional antibiotic agents were pivotal in creating this quandary, and they represent an infeasible solution to this problem, especially in terms of the time and cost associated with developing structurally novel drugs distinct enough from current therapies to provide a substantive improvement to the status quo.
Conceptual Advantages to an Anti-Sense Approach
There is a recent and concerted research effort toward the development of newer classes of antibiotic agents that self-consciously have a microbiome-sparing effect. To achieve this effect, these new treatments have a completely different approach to bacterial killing, one targeting the central dogma of molecular biology by disrupting essential bacterial processes with the goal of genus or even species level specificity. One of these approaches is referred to broadly as an anti-sense approach, with anti-sense referencing the complementarity between two strands of nucleic acids capable of annealing one to another.
This approach seeks to use oligonucleotides to disrupt the expression of key bacterial genes; although various approaches are available, this most often involves the complementary binding of a modified anti-sense oligonucleotide (ASO) to a specific messenger ribonucleic acid (RNA) molecule (mRNA). Once the ASO binds to its mRNA target (Fig. 1a and 1b), assembly of ribosomal subunits on the mRNA is prohibited through a process referred to as steric hindrance, a category of intermolecular interactions where the physical bulk provided by the ASO prevents certain physiochemical reactions from taking place. In this scenario, translation is typically prevented by the ASO binding to the mRNA transcript near the Shine-Dalgarno and start codon sequences (pivotal locations for ribosomal assembly and initiation of translation), preventing ribosomal assembly and thus thwarting the production of a protein, with the resultant loss of a bacterial function.
FIG. 1.
Assembly of the small (30S) and large (50S) ribosomal subunits is a key initiation step in messenger ribonucleic acid (mRNA) translation in bacteria. The presence of an anti-sense oligonucleotide complementary to these key sites in mRNA can prevent ribosomal subunit assembly, thus preventing translation.
This approach has several potential advantages compared with currently available antibiotic agents. First, while certain gene pathways may be advantageous enough to be conserved among bacteria of different taxonomic levels, the ability to create an ASO specifically targeting the mRNA of a particular gene in a particular genus or species of bacterium provides a therapy with much more specificity than is available currently. This would not only decrease the incidence of antibiotic-associated diarrhea, but it would decrease the incidence of CDI normally associated with antibiotic agents. As the length of the ASO increases, so does the specificity of the ASO for its target mRNA, and so does the strength of ASO binding to its target mRNA, ostensibly contributing to improved specificity and treatment efficacy, respectively.
Second, while genetic variation of targeted genes is expected to occur, a simple change to the sequence of the ASO addresses this form of AMR; ASOs are relatively inexpensive, allowing for time and cost-feasible flexibility to acquired genetic mutations. Third, ASOs provide a wider range of potential therapeutic targets. Using CDI as an example, the ability to decrease the volume of toxin production could lessen the severity of colitis symptoms. As a toxin-dependent disease, this approach would prevent the development of life-threatening forms of CDI, obviating the need for surgical intervention by rendering the infection closer to an asymptomatic carrier state. The ability to disrupt sporulation pathways, either preventing spore formation or preventing spore germination, would address the major disease reservoir of this and many other infections.
Challenges to an Anti-Sense Approach
Several difficulties must be overcome for any anti-sense approach to be successful. The first is to identify a bacterial gene pathway that, if interrupted, would produce a measurable benefit in the infected human host. Candidate pathways typically involve bacterial growth, deoxyribonucleic acid (DNA)/RNA polymerases, and lipid metabolism [13]. Once a gene pathway is selected, studying the secondary structure of target RNA molecules using in silico techniques is performed to identify the sequence and the region of RNA where steric hindrance of ribosomal assembly will be greatest. This will also predict the sequences that will offer the most favorable thermodynamics for ASO-RNA binding. In addition, the in silico process also involves evaluating for off-target ASO hybridization, which can be computationally predicted with a high degree of certainty.
Once a sequence of ASO is selected, the next step is selecting a type of ASO; although a detailed description is beyond the purpose of this article, native oligonucleotides would generally be a poor choice because of their sensitivity to nuclease degradation. Therefore, anti-sense approaches use modified oligonucleotides generally 10–30 bases in length, with various modifications involving the phosphate group, the sugar moiety, or the form of linkage between bases (a detailed review by Hegarty and Stewart [14] describes the relative advantages of these modified nucleic acids). These modifications allow a stronger ASO-target complexation, while rendering these ASOs more or completely resistant to nucleases.
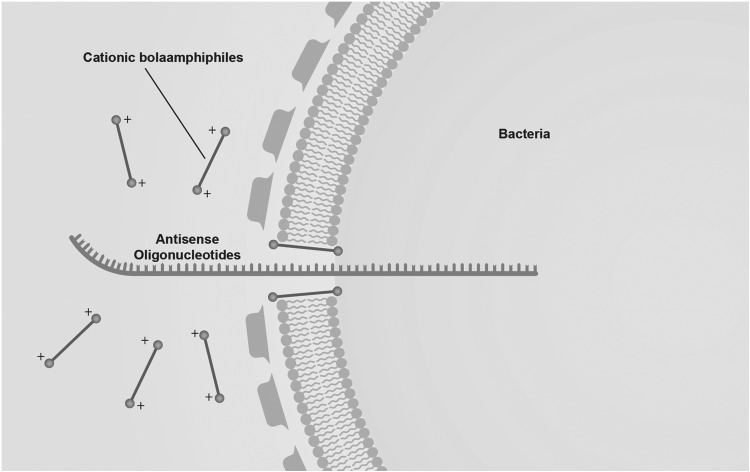
The next consideration is what delivery strategy will be selected. Bacterial uptake of free ASOs is poor, because of a combination of electrostatic forces, barriers to diffusion through the extracellular environment, and size mismatches related to the cell wall. As opposed to eukaryotic cells, there are fewer validated delivery systems for ASOs in prokaryotes. Typically, either cell-penetrating peptides or nanomaterials composed of lipids, inorganic compounds, or various polymers have been described [14,15]. The delivery system must (1) complex with the ASO to be delivered, while (2) concentrating enough ASOs to deliver an effective dose, and (3) protecting ASOs from degradation in the extracellular environment (Fig. 2). The delivery system must also avoid exerting any antibacterial effects in order to maintain the preservation of the untargeted gut biomass, while also posing no toxicity to host tissue such as the alimentary tract.
FIG. 2.
Nanocarriers are required to introduce effective amounts of anti-sense oligonucleotide into bacteria. One approach uses cationic bolaamphiphiles, which are thought to create transient pores in bacterial membranes because of electrostatic interactions with the negatively charged phospholipid cardiolipin.
Evidence for Efficacy of Anti-Sense Therapies
The majority of research with anti-sense therapies has focused on the use of phosphorodiamidate morpholino oligomers (PMO) conjugated to cell penetrating peptides (CPP-PMO). One of the first reports [16] using in vivo modeling with mice described the successful targeting of the acpP gene (acyl carrier protein involved in fatty acid biosynthesis) in Escherichia coli using an 11 base PMO, demonstrating a 10-fold sustained reduction in bacterial growth at 6, 12, and 24-hour time points after exposure. Promising in vivo data have also been produced targeting Pseudomonas aeruginosa.
In a study [17] from 2017, CPP-PMOs targeted acpP, lpxC (involved in the deacetylation process of lipid A synthesis), and rpsJ (involved in producing the 30S ribosomal protein S10) genes. Not only was PMO therapy effective in a mouse model, but also combinations of PMOs as well as combining PMOs with conventional antibiotic agents demonstrated synergistic effects beyond single PMO use alone. Interestingly, PMO therapy targeting the acpP gene was effective at both preventing as well as reducing existing biofilm layers. In vivo data [18] using mice also suggests the effectiveness of CPP-PMO anti-sense therapies eradicating Acinetobacter lwoffii and Acinetobacter baumannii, with minimum inhibitory concentrations (MICs) of PMOs being comparable to MICs for known conventional antibiotic agents, with the additional finding of decreased inflammation attributable to bacteria.
Further, Daly et al. [19] described the use of CPP-PMOs targeting the MCR-1 (mobilized colistin resistance) gene, where anti-sense therapies restored sensitivity of E. coli to colistin; these data demonstrate the potential expanded arsenal provided by such molecular therapies that can either target bacteria directly or that can silence AMR mechanisms allowing the concomitant use of conventional antibiotic agents.
There has been recent investigation into applying anti-sense therapies to the management of CDI. Given the degree to which gut dysbiotic states participate in the pathogenesis of CDI, including potentially contributing to recurrent forms of this infection [20], anti-sense therapies offer a conceptually appealing alternative to conventional drugs because of their potential for greater organismal specificity. Although there are no current in vivo data, there are recent in vitro data using cell cultures to suggest a proof-of-principle to this approach for CDI.
The first study in the literature on anti-sense approaches in CDI, published in 2016 [21], tested a group of 2'-O-methyl phosphorothioate gapmer ASOs targeting any of five essential C. difficile genes. The selection of nanocarrier to introduce ASO into bacteria in this study is intriguing, borrowing from principles of mitochondrial medicine. The inner mitochondrial membrane is the one eukaryotic membrane enriched by cardiolipin (CL), a phospholipid with surfactant properties [22] and one that aggregates in areas of membranes with greater angulation, playing a pivotal role in the development of structures such as septa. The CL is also a common component of bacterial membranes, with the similarity between mitochondrial and bacterial plasmalemmae reflecting a shared origin as related to endosymbiont bacteria occupying an intracellular position and eventually transitioning from an independent organism to an organelle.
In this study, cationic vesicles (bolasomes) composed of dequalinium chloride previously used to deliver small molecules and plasmid DNA across the inner mitochondrial membrane, were re-purposed to deliver ASO gapmers into gapmer, with four of the tested gene targets achieving nanomolar minimum inhibitory concentrations against C. difficile. The best performing ASOs targeted polymerase genes rpoB and dnaE.
Because dequalinium is known to have an antibiotic effect at higher concentrations, a second study [23] on anti-sense therapy for CDI focused on retaining the best performing ASO, but now tested with one of three novel nanocarriers designed using structure-activity relationship analyses to create a carrier with no antibacterial activity and with no toxicity to colonocytes. In this more recent study, nanocarriers without ASO demonstrated little antibacterial activity against bacterial cultures of C. difficile, E. coli, Bacteroides fragilis, and Enterococcus faecalis, with limited colonocyte toxicity using Caco-2 cells, all at concentrations capable of delivering an effective concentration of ASO. The best performing nanocarriers loaded with ASO demonstrated a clinically relevant MIC90 to C. difficile of 19 mcM.
Challenges and Future Directions
Compared with eukaryotic systems, the translational machinery of prokaryotes is “simpler” to disrupt. Unlike eukaryotic systems, however, prokaryotic cells have barriers to oligonucleotide penetration (cell walls, outer membranes) in addition to a plasma membrane. These barriers are relatively understudied from the standpoint of drug delivery, and in the case of plasma membranes, their surface chemistry is far more complex than previously thought, with myriad microdomains that will need to be exploited for anti-sense approaches.
Drug delivery into the bacterial cytoplasm requires a balance between exploiting features of bacteria that are conserved enough across genera to make the nanocarrier useful, while not producing an antibiotic effect. While the nanocarrier should be able to deliver drug into any targeted bacteria, the specificity of the drug depends on the ASO. Many of the most important gene pathways have redundancy that would require targeting more than one gene to interrupt a single bacterial function. The more important a gene pathway, the larger the copy number of RNA transcripts, requiring a larger number of ASOs to effectively silence a gene. Delivery of the drug complex can be of relative concern if delivered by mouth, both in terms of delivering an effective dose to the affected segment of intestine and in terms of avoiding enzymatic degradation.
In the anti-sense antimicrobial literature, there is a large number of publications based on cell culture experiments, with a lesser amount of data using animal models. This speaks to the convergence of challenges with this approach in terms of cost of development of drugs for research purposes (less expensive than conventional drugs, but still expensive for design in research laboratories), as well as challenges in terms of drug delivery and effectiveness. During the past five years, the number of researchers in this arena has increased significantly, which may promulgate forward this approach to antibacterial therapy.
The one overarching benefit of an anti-sense approach is that of molecular precision—choosing bacteria based on unique, differentiating molecular features, and further selecting a precise function to disrupt. Anti-sense therapies apply the central dogma of molecular biology toward the concept that for any encoded bacterial function, the same function can be disrupted.
Author Disclosure Statement
This manuscript was supported (PI – Stewart) by funding from the National Institutes of Allergy and Infectious Diseases (2 R21 AI132353-01).
References
- 1. Centers for Disease Control and Prevention. Antibiotic/antimicrobial resistance. www.cdc.gov/drugresistance/index.html (Last accessed July16, 2018)
- 2. World Health Organization. Antimicrobial resistance. www.who.int/antimicrobial-resistance/en/ (Last accessed on July16, 2018)
- 3. Thorpe KE, Joski P, Johnston KJ. Antibiotic-resistant infection treatment costs have doubled since 2002, now exceeding $2 billion annually. Health Aff (Millwood) 2018;37:662–669 [DOI] [PubMed] [Google Scholar]
- 4. Conly JM, Johnston BL. Where are all the new antibiotics? The new antibiotic paradox. Can J Infect Dis Med Microbiol 2005;16:159–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Munita JM, Arias CA. Mechanisms of antibiotic resistance. Microbiol Spectr 2016;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Woodford N, Ellington MJ. The emergence of antibiotic resistance by mutation. Clin Microbiol Infect 2007;13:5–18 [DOI] [PubMed] [Google Scholar]
- 7. Barlow M. What antimicrobial resistance has taught us about horizontal gene transfer. Methods Mol Biol 2009;532:397–411 [DOI] [PubMed] [Google Scholar]
- 8. Baur D, Gladstone BP, Burkert F, et al. . Effect of antibiotic stewardship on the incidence of infection and colonisation with antibiotic-resistant bacteria and Clostridium difficile infection: A systematic review and meta-analysis. Lancet Infect Dis 2017;17:990–1001 [DOI] [PubMed] [Google Scholar]
- 9. Brown K, Valenta K, Fisman D, et al. . Hospital ward antibiotic prescribing and the risks of Clostridium difficile infection. JAMA Intern Med 2015;175:626–633 [DOI] [PubMed] [Google Scholar]
- 10. Kaplan JB. Antibiotic-induced biofilm formation. Int J Artif Organs 2011;34:737–751 [DOI] [PubMed] [Google Scholar]
- 11. Gil F, Lagos-Moraga S, Calderón-Romero P, et al. . Updates on Clostridium difficile spore biology. Anaerobe 2017;45:3–9 [DOI] [PubMed] [Google Scholar]
- 12. Gerber M, Walch C, Löffler B, et al. . Effect of sub-MIC concentrations of metronidazole, vancomycin, clindamycin and linezolid on toxin gene transcription and production in Clostridium difficile. J Med Microbiol 2008;57:776–783 [DOI] [PubMed] [Google Scholar]
- 13. Rasmussen LC, Sperling-Petersen HU, Mortensen KK. Hitting bacteria at the heart of the central dogma: Sequence-specific inhibition. Microb Cell Fact 2007;6:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hegarty JP, Stewart DB Sr. Advances in therapeutic bacterial anti-sense biotechnology. Appl Microbiol Biotechnol 2018;102:1055–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ahmed M. Peptides, polypeptides and peptide-polymer hybrids as nucleic acid carriers. Biomater Sci 2017;5:2188–2211 [DOI] [PubMed] [Google Scholar]
- 16. Geller BL, Deere J, Tilley L, Iversen PL. Antisense phosphorodiamidate morpholino oligomer inhibits viability of Escherichia coli in pure culture and in mouse peritonitis. J Antimicrob Chemother 2005;55:983–988 [DOI] [PubMed] [Google Scholar]
- 17. Howard JJ, Sturge CR, Moustafa DA, et al. . Inhibition of Pseudomonas aeruginosa by peptide-conjugated phosphorodiamidate morpholino oligomers. Antimicrob Agents Chemother 2017;24:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Geller BL, Marshall-Batty K, Schnell FJ, et al. . Gene-silencing anti-sense oligomers inhibit Acinetobacter growth in vitro and in vivo. J Infect Dis 2013;208:1553–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Daly SM, Sturge CR, Felder-Scott CF, et al. . MCR-1 Inhibition with peptide-conjugated phosphorodiamidate morpholino oligomers restores sensitivity to polymyxin in Escherichia coli. Mbio 2017;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lamendella R, Wright JR, Hackman J, et al. . Antibiotic treatments for Clostridium difficile infection are associated with distinct bacterial and fungal community structures. mSphere 2018;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hegarty JP, Krzeminski J, Sharma AK, et al. . Bolaamphiphile-based nanocomplex delivery of phosphorothioate gapmer anti-sense oligonucleotides as a treatment for Clostridium difficile. Int J Nanomedicine 2016;11:3607–3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mileykovskaya E, Dowhan W. Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim Biophys Acta 2009;1788:2084–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sharma AK, Krzeminski J, Weissig V, et al. . Cationic amphiphilic bolaamphiphile-based delivery of anti-sense oligonucleotides provides a potentially microbiome sparing treatment for C. difficile. J Antibiot (Tokyo) 2018l71:713–721 [DOI] [PMC free article] [PubMed] [Google Scholar]