Abstract
Stem cell cultures within perfusion bioreactors, while efficient in obtaining cell numbers, often lack the similarity to native tissues and consequently cell phenotype. We develop a three-dimensional (3D)-printed fluidic chamber for dynamic stem cell culture, with emphasis on control over flow and substrate curvature in a 3D environment, two physiologic features of native tissues. The chamber geometry, consisting of an array of vertical cylindrical pillars, facilitates actin-mediated localization of human mesenchymal stem cells (hMSCs) within ∼200 μm distance from the pillars, enabling spatial patterning of hMSCs and endothelial cells in cocultures and subsequent modulation of calcium signaling between these two essential cell types in the bone marrow microenvironment. Flow-enhanced osteogenic differentiation of hMSCs in growth media imposes spatial variations of alkaline phosphatase expression, which positively correlates with local shear stress. Proliferation of hMSCs is maintained within the chamber, exceeding the cell expansion in conventional static culture. The capability to manipulate cell spatial patterning, differentiation, and 3D tissue formation through geometry and flow demonstrates the culture chamber's relevant chemomechanical cues in stem cell microenvironments, thus providing an easy-to-implement tool to study interactions among substrate curvature, shear stress, and intracellular actin machinery in the tissue-engineered construct.
Keywords: : fluidic, perfusion culture, 3D printing, mesenchymal stem cells
Introduction
During tissue formation, complex cellular activities such as proliferation and differentiation are regulated through distance-dependent signaling events, creating the need to manipulate cell position in a tissue to modulate signaling. The ability to pattern cells within engineered platforms therefore permits control of cell–cell interactions, allowing generation of in vitro models of tissues, organoids, and subsequent relevant mechanistic cellular studies. To create relevant stem cell niche-like microenvironments, efforts have been made to form three-dimensional (3D) geometries of artificial tissues inside perfusion systems,1,2 which more closely mimic natural tissues than cells in static two-dimensional (2D) cultures, thus displaying physiologically relevant cell phenotypes.3
Perfusion bioreactors aid in creating physiologic stem cell microenvironment through shear stress on the cell surface, as well as media and oxygen distribution, resulting in improved cell seeding efficiency,4–6 cell proliferation,7–10 and osteogenic differentiation of mesenchymal stem cells (MSCs).11–20 Integration of 3D culture and cell patterning capability into dynamic perfusion systems for cell cultures will aid in the development of tissue models with relevant physiological stem cell environments, for studies of chemomechanical responses of cells, as well as possible ex vivo expansion of cells. Our goal is to create a cell culture platform that allows the creation of a model stem cell microenvironment through spatial patterning of cells, which can be used to study interactions of key cells of the bone marrow microenvironment, that is, MSCs, osteoblasts, and endothelial cells (ECs), enabling new insights into stem cell biology.
To facilitate cell spatial patterning, precisely controlled substrate geometry within culture systems allows tailoring of the number of cells per unit area or volume, cell–cell distance, and flow pattern, which might modulate key cell–cell signaling in the formed tissue. Complex 3D geometries, however, introduce numerous parameters that influence stem cell behavior, for example, curvature21–23 and complex flow patterns.24–26 Therefore, understanding the contribution of these parameters to cell adhesion, proliferation, and differentiation is crucial for designing more effective culture system.
Such studies are possible in fluidic channels, which can provide spatial and temporal control of cell growth and stimuli through substrate geometry and fluid transport, while simultaneously providing a platform for cell imaging, image-based analysis, and further biochemical analysis of single cells in tissues27; therefore, a fluidic system remains as our base platform for this study. Existing fluidic platforms to support 3D cell culture have been reported, however, the three-dimensionality is typically achieved through cell encapsulation in scaffolds,28–31 after which the cell culture is placed in a perfusion system.32–36 The novelty of our fluidic culture system is the incorporation of cellular patterning simply through substrate curvature and flow-driven shear stress in a scaffold-free fluidic design to form a 3D complex tissue.
By combining the advantages of shear stress from flow perfusion, precise geometrical features from 3D printing (3DP), and image-based analysis capability of a fluidic system, we aim to engineer and characterize the model stem cell environment created inside the fluidic culture chamber. Our culture chamber involves an array of vertical cylindrical pillars, which provides additional surface for cells to grow on while obtaining beneficial shear stress due to the media flow. Further tuning of the pillar-to-pillar distance enables formation of 3D human mesenchymal stem cell (hMSC) culture simply from initially 2D seeded cells, without the presence of external supporting scaffolds, as well as spatial control of cell locations. Such features allow for culture and creation of a tissue structure within the stem cell microenvironment with several controllable features, including shear stress, presence of spatially controlled multiple cell types, and three-dimensionality.37
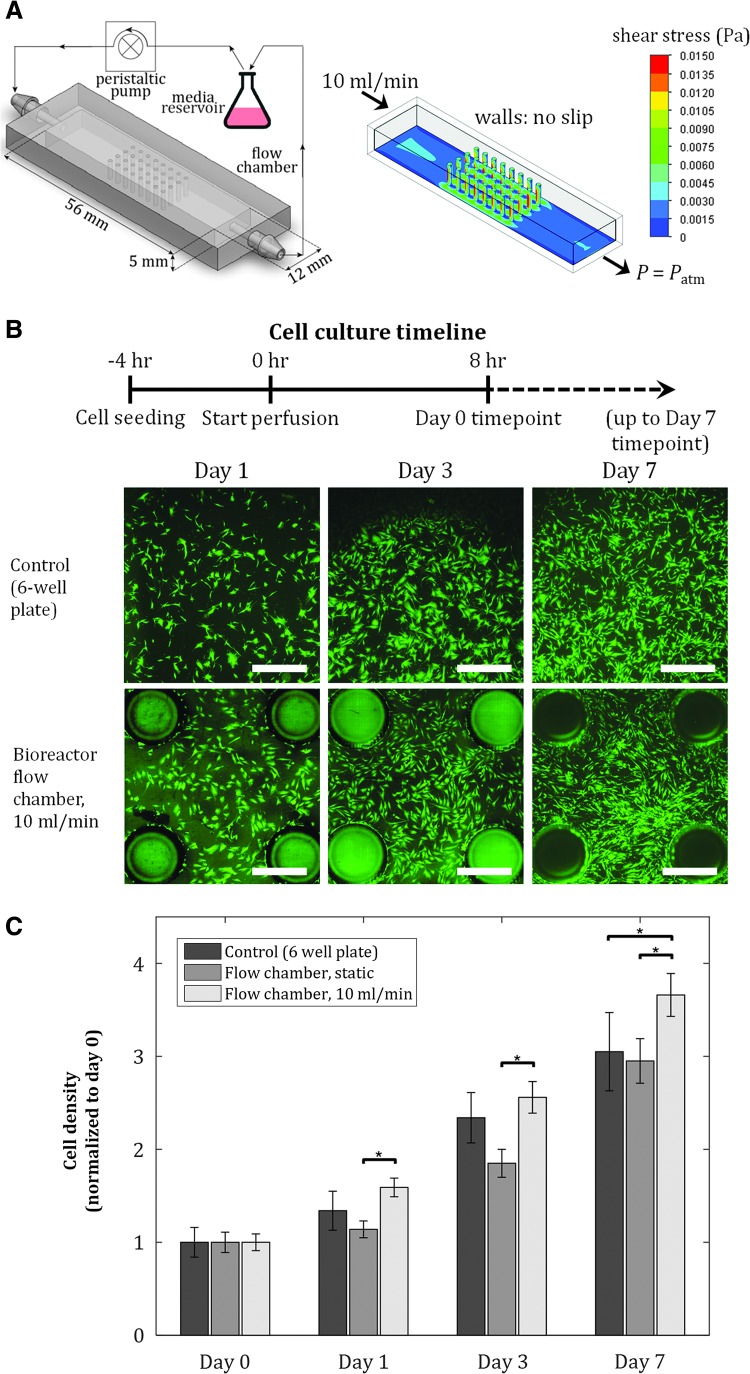
The fluidic culture chamber is fabricated using 3DP technology, allowing precisely controlled geometry through computer-aided design (CAD). In addition, this fluidic culture system allows image-based analysis of the cells within the chamber at any time point during growth or differentiation, enabling quantification of correlations between locally observed cellular responses and precisely tuned flow patterns. The chambers are then integrated with off-the-shelf tubing and a peristaltic pump to form a continuous fluidic perfusion culture system (Fig. 1A). We hypothesize that the environment created in this culture platform can provide spatial control of hMSC growth and differentiation (to mimic site-specific cellular processes) due to local flow patterns and surface curvatures, in addition to enhancement of osteogenic differentiation. Understanding cellular responses to flow and geometrical cues in our fluidic culture platform allows us to modify the design for diverse stem cell applications.
FIG. 1.
Proliferation of hMSCs is maintained in fluidic culture chambers. Initial seeding density = 40 cells/mm2. (A) Schematic of experimental setup and culture chamber (left), and shear stress distribution on chamber inner surfaces from fluid flow simulation (right). Pillar diameter: 1 mm, pillar-to-pillar distance: 2.5 mm. (B) Fluorescent images of hMSCs at various time points for control, static, and dynamic conditions in culture chambers. Scale bar = 1 mm. (C) Cell proliferation after 7 days is maintained in dynamic culture conditions, achieving at least the same cell expansion as in two-dimensional static cultures. Values are averages of four experiments and error bars are standard deviations (*p ≤ 0.05). hMSCs, human mesenchymal stem cells.
Based on these advantages, we use our fluidic culture chamber to dynamically culture hMSCs; specifically, the objectives of this study are to (1) quantify the growth and osteogenic differentiation of hMSCs in our culture chamber, (2) utilize local flow to pattern alkaline phosphatase (ALP) expression in hMSCs, and (3) utilize substrate curvature to spatially pattern cocultures of hMSCs and ECs, two critical cells in the bone marrow microenvironment. We maintain hMSC culture under dynamic conditions in our fluidic chamber. Flow enhances osteogenic differentiation, as quantified by the increased expression of the early osteogenic marker ALP, indicating presence of osteoblasts in culture, another key cell of the bone marrow environment.38 In addition, we utilize the pillar geometry inside the culture chamber to create 3D structure of hMSCs, as well as spatially patterned cocultures of hMSCs and ECs. The ability of our culture system to allow hMSC growth, differentiation to osteoblasts, spatial patterning of hMSCs and ECs in cocultures, as well as 3D tissue growth, allows the fabrication of custom-tailored tissue models for studies of chemomechanical cellular behavior in a controlled stem cell microenvironment.
Materials and Methods
Fabrication and treatment of culture chambers
Culture chambers were designed in SolidWorks (Dassault Systèmes, Vélizy-Villacoublay, France) and fabricated using the EnvisionTEC Perfactory 3D printer, with EShell® 300 as the printing material. Following printing, chambers were washed in isopropanol for 1 h to eliminate residual, uncross-linked material, and then flash cured. For sterilization, chambers were filled with ethanol and then exposed to ultraviolet light for 30 min. Chambers were then rehydrated by serial washes in the following sterile solutions: (1) 75% ethanol/25% phosphate-buffered saline (PBS), (2) 50% ethanol/50% PBS, (3) 25% ethanol/75% PBS, and (4) 100% PBS, then stored in PBS. Before cell seeding, flow chambers were coated with 3 μg/cm2 fibronectin (Sigma-Aldrich, St Louis, MO) in PBS for 1 h at 37°C to facilitate cell attachment.
Cell culture and seeding
hMSCs (RoosterBio, Frederick, MD) were cultured in the accompanying High Performance Media Kit (RoosterBio). For cell seeding, hMSCs were harvested at passage 3, suspended in growth media, that is, high-glucose Dulbecco's modified Eagle's medium containing l-glutamine (Gibco, Grand Island, NY), supplemented with 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA), 1% v/v penicillin/streptomycin (Gibco), and 0.1 mM nonessential amino acids (Invitrogen), and plated in six-well plates or seeded into culture chambers at density of 40 cells/mm2 chamber-bottom surface area. For osteogenic differentiation, hMSCs were cultured in growth media supplemented with 100 nM dexamethasone (Sigma-Aldrich), 173 mM ascorbic acid (Sigma-Aldrich), and 10 mM β-glycerophosphate (Sigma-Aldrich). Human umbilical vein endothelial cells (HUVECs; Lonza, Walkersville, MD) were cultured in EC growth media (Lonza). Jurkat cells (ATCC, Rockville, MD) were cultured in RPMI-1640 medium (ATCC) supplemented with 10% FBS. All cells were cultured at 37°C with 5% CO2. For hMSC-EC coculture experiments, hMSCs were first seeded into the chambers and allowed to adhere for 12 h under static conditions, and then, HUVECs were seeded and allowed to adhere for 12 h. For hMSC-Jurkat coculture experiments, hMSCs were seeded into the chambers and allowed to adhere for 12 h, then Jurkat cell suspension in media (density: 105 cells/mL) were circulated through the chambers. For actin inhibition experiments, actin fibers in hMSCs or HUVECs were inhibited by treating cells in suspension with 10 μM cytochalasin-D (Sigma-Aldrich) in PBS at 37°C for 1 h before seeding.
Static and dynamic culture
Three experimental groups were tested: static culture in six-well plates (control), static culture in fluidic chambers, and dynamic culture in fluidic chambers. After seeding, cells were allowed to adhere to the culture chambers or six-well plates for 4 h. Cells in the static six-well plates and chambers were cultured in 2 mL of media per well or per chamber. For the dynamic group, the culture chambers were connected to a media reservoir through silicone tubing, inner diameter 0.8 mm (Cole Parmer, Chicago, IL), then connected to an L/S Multichannel Pump System (Cole Parmer) to form the fully integrated perfusion system assembly. Flow was then driven by the peristaltic pump at a flow rate of 10 mL/min. All groups were cultured at 37°C with 5% CO2, and medium was changed every 3 days.
RNA extraction and reverse transcription
Following static and dynamic culture for 7 days, cells from each culture were harvested using trypsin/0.25% EDTA (Life Technologies, Grand Island, NY). A cell pellet was formed by centrifugation and washed with PBS two times. RNA isolation was performed using the RNeasy Plus Mini Kit (Qiagen, Germantown, MD) following standard manufacturer's protocols. Isolated RNA from each sample was then reverse transcribed to cDNA using a High Capacity cDNA Archive Kit (Life Technologies).
Real-time quantitative polymerase chain reaction
Real-time quantitative polymerase chain reaction (RT-qPCR) was performed by combining the cDNA solution with a Universal Master Mix (Life Technologies), along with oligonucleotide primers and Taqman probes for ALP, and compared to the endogenous control gene, glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Life Technologies). The reaction was performed using a 7900HT real-time PCR System (Applied Biosystems, Foster City, CA) at thermal conditions of 2 min at 50°C, 10 min at 95°C, 40 cycles of 15 s at 95°C, and 1 min at 60°C. The relative gene expression level of each target gene was then normalized to the mean of the GAPDH in each group.
Cell viability assay and counting
Cell viability was assessed using a fluorescent Live/Dead assay (Invitrogen) following standard protocols. The following solutions were prepared: 4 μM Calcein-AM (Invitrogen) and 4 μM ethidium homodimer (Invitrogen) in Hank's Balanced Salt Solution (HBSS; ThermoFisher Scientific), delivered into the chambers or six-well plates containing cells, and incubated for 45 min. Samples were then washed with HBSS before imaging. Fluorescent images were then taken at three predetermined regions for each sample using a fluorescence microscope (Axiovert 40 CFL; Zeiss, Jena, Germany) equipped with a digital camera (11.2 Color Mosaic; Diagnostic Instruments, Sterling Heights, MI). This process was repeated for all the four samples in each group. Cell counting was performed on each fluorescent image of live cells and averaged over all samples in a group (n = 4).
Cell staining and immunofluorescence imaging
To visualize ALP expression, cells were fixed in 4% paraformaldehyde for 30 min at room temperature. Following fixation, cells were washed with PBS three times, permeabilized with PBS containing 1% bovine serum albumin (BSA), 10% goat serum, 0.3 M glycine, and 0.1% tween for 5 min, and blocked with 1% BSA in PBS for 30 min at 37°C. Samples were then incubated with anti-ALP primary monoclonal antibody (Abcam, Cambridge, MA) at 1:100 dilution overnight at room temperature. To visualize the proteins, samples were then incubated in Alexa Fluor® 555 secondary antibody at 1:500 dilution for 1 h at room temperature. F-actin was stained using rhodamine phalloidin (Life Technologies) and DNA was stained with DAPI (Life Technologies). To visualize hMSCs and ECs in coculture, hMSCs and HUVECs were transduced with BacMam GFP Transduction Control and CellLight® Nucleus-RFP (ThermoFisher Scientific), respectively, before seeding. Cells were imaged using a Leica SP5X confocal microscope (Leica Microsystems, Wetzlar, Germany), and subsequently analyzed using ImageJ (National Institutes of Health, Bethesda, MD) and MATLAB (The MathWorks, Inc., Natick, MA).
Intracellular Ca2+ imaging and analysis
Before Ca2+ imaging, Fluo-4 calcium dye (Invitrogen) was administered to cells in growth medium and left to incubate for 30 min. Fluorescence from the calcium was detected using an argon laser at 488 nm. For each sample, as 1 mL of 10 μM ATP (Sigma-Aldrich) was delivered into the cell culture, a movie was taken at a single focal plane for 25 min at a rate of 1 frame/s. Subsequent image analysis and data processing were performed in MATLAB.39,40 In brief, during the cells response to ATP, each cell's fluorescent intensity I(t) was obtained by averaging the pixel color values of 25 pixels around each cell's center of mass, then normalized to a baseline intensity Ir to obtain the response curve R(t), defined as R(t) = [I(t) − Ir]/Ir. A cell was referred to as responsive if, at any time, R(t) ≥ 0.5. The oscillation peaks were next detected in MATLAB using the function peakfinder.m (available through MATLAB File Exchange). If a cell's response had two or more peaks, we referred to the cell as a responsive, oscillating cell (details in Supplementary Materials section; Supplementary Data are available online at www.liebertpub.com/tea).
Fluid flow simulations
To quantify shear stress in the culture chambers, flow simulations were performed in SolidWorks. Inlet flow rate of 10 mL/min was applied with a fully developed flow behavior. Outlet condition was set to open to atmospheric pressure, and all walls were no-slip boundaries. The fluid had thermodynamic properties of water at atmospheric pressure and 37°C.
Statistical analyses
Each sample was analyzed in triplicate. In each figure, we reported mean values of samples, error bars which are standard deviations of the means, as well as relevant statistical relationships. Statistical significance between groups was determined by performing a two-sample t-test. Confidence intervals of 95% (α = 0.05), 99% (α = 0.01), and 99.9% (α = 0.001) were used for all analyses.
Results
Proliferative capacity of hMSCs is maintained in dynamic culture condition in fluidic culture chambers
We fabricated culture chambers with dimensions shown in Figure 1A, pillar diameter of 1 mm, pillar-to-pillar distance of 2.5 mm, and total surface area of 1140 mm2, compared to 900 mm2 surface area of a single well in a six-well plate culture dish. To avoid entrance effects and to allow for fully developed laminar flow, an inlet length of 19 mm was provided in the chambers before exposure to the pillars (Fig. 1A); this is significantly larger than the minimum entrance length for a flow rate of 10 mL/min in this geometry, thus satisfying the conditions for laminar flow in a noncircular pipe.41
Using flow simulations, the shear stress profile inside the flow chambers was quantified (Fig. 1A). Shear stress of up to 15 mPa was observed, which is significantly below the reported shear stress value that causes mechanical damage and detachment of MSCs.42,43 We quantified the areal density of cells in the culture chambers under static and dynamic conditions (10 mL/min), and in six-well plates (control), at four time points postseeding: day 0 (8 h after flow is turned on), day 1, 3, and 7 (Fig. 1B). The areal density at each time point was normalized by the corresponding cell density at day 0 (Fig. 1C). At each time point, the density of hMSCs in static condition in the chambers was not statistically different from that of the control group, while dynamic conditions in the chambers achieved modestly higher cell density at day 7 than the cell density in the static groups. Overall, hMSC proliferation in the dynamic culture chamber was maintained, achieving at least the same cell expansion as in 2D static cultures, demonstrating the culture chamber's conducive environment for hMSC culture.
Flow-induced shear stress on cells creates spatially patterned enhancement of osteogenic differentiation
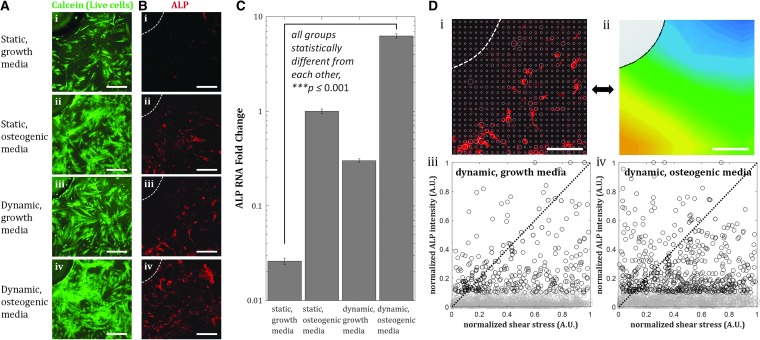
Osteogenic differentiation of hMSCs has been reported to be enhanced by shear stress, as shown by increased expression of osteogenic markers ALP, PGE2, OPN, OC, COX-2, RunX2, Col1, and mineralized matrix production with shear stresses in the range of O(10−4) Pa to O(1) Pa.44,45 We performed an osteogenic differentiation study on hMSCs cultured inside the culture chambers under static and dynamic conditions for 7 days, in either growth or osteogenic differentiation media. We performed immunostaining for ALP, an early osteogenic marker in hMSCs, and quantified the relative expression of ALP using quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR). Immunofluorescence images revealed enhanced expression of ALP in dynamic culture compared to static conditions, in both growth and osteogenic media (Fig. 2B). qRT-PCR results confirmed these observations, where 10-fold enhancement of ALP expression by flow was observed (Fig. 2C).
FIG. 2.
Shear stress enhances osteogenic differentiation after 7 days in culture chambers. (A) Fluorescent images of hMSCs near pillars inside flow chambers after 7 days of culture. (B) Immunofluorescent images of ALP expression in hMSCs in chambers under static (i, ii) and dynamic condition, flow rate = 10 mL/min (iii, iv). Dashed white lines indicate the edge of a pillar. Scale bar = 200 μm. (C) ALP expression in growth and osteogenic media is enhanced by shear stress. Up to ∼10-fold enhancement is achieved for the dynamic condition in growth media. Values are averages of three experiments and error bars are standard deviations (***p ≤ 0.001). (D) Analysis of local values of normalized ALP expression (i) and local shear stress (ii), and the correlation of the two images. Scale bar = 200 μm. Normalized ALP intensity versus normalized shear stress plots illustrate the correlation between the two quantities for growth media (iii) and osteogenic media (iv). Dashed black line with slope = 1 indicates perfect positive correlation. Points in the dynamic, osteogenic media group are scattered further away from the dashed line, with points corresponding to high ALP expression near zero-shear stress regions, meaning smaller correlation coefficient between ALP expression and shear stress. ALP, alkaline phosphatase.
We also observed spatial variations of ALP expression in the case of flow-enhanced osteogenic differentiation in growth media. When we quantified the normalized expression of ALP, 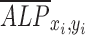 , at various distances from the pillars (details in Supplementary Materials section), we observed the most pronounced expression at the distance of ∼600 μm from the pillar (Supplementary Fig. S1B, E), while the normalized ALP expression in the dynamic group with osteogenic media was observed in all distances (Supplementary Fig. S1A, D). We also calculated the Pearson correlation coefficient
, at various distances from the pillars (details in Supplementary Materials section), we observed the most pronounced expression at the distance of ∼600 μm from the pillar (Supplementary Fig. S1B, E), while the normalized ALP expression in the dynamic group with osteogenic media was observed in all distances (Supplementary Fig. S1A, D). We also calculated the Pearson correlation coefficient 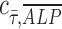 to quantify the correlation between ALP expression and shear stress in the two dynamic groups as defined in Equation (1):
to quantify the correlation between ALP expression and shear stress in the two dynamic groups as defined in Equation (1):
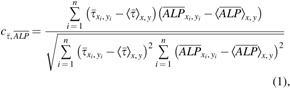
where 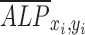 is the normalized ALP intensity at location (xi, yi),
is the normalized ALP intensity at location (xi, yi), 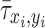 is the normalized shear stress at location (xi, yi),
is the normalized shear stress at location (xi, yi), 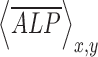 is the average value of
is the average value of 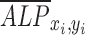 ,
, 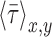 is the average value of
is the average value of 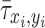 , and n is the number of measurements taken from an image, as indicated by the white open circles in Figure 2D-i. Note that a value of
, and n is the number of measurements taken from an image, as indicated by the white open circles in Figure 2D-i. Note that a value of 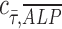 = 1 implies positive correlation between the two quantities,
= 1 implies positive correlation between the two quantities, 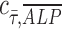 = −1 implies a negative correlation, and
= −1 implies a negative correlation, and 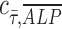 = 0 implies no correlation. Flow-enhanced ALP expression in the growth media revealed
= 0 implies no correlation. Flow-enhanced ALP expression in the growth media revealed 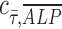 = 0.24, while this dynamic condition in osteogenic media resulted in
= 0.24, while this dynamic condition in osteogenic media resulted in 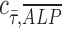 = −0.07. This suggests that shear stress weakly correlates with ALP expression in dynamic condition with growth media; however, in osteogenic differentiation media, the effect of shear stress is less prominent than that in growth media, therefore resulting in correlation coefficient close to zero. In addition, a perfect positive correlation
= −0.07. This suggests that shear stress weakly correlates with ALP expression in dynamic condition with growth media; however, in osteogenic differentiation media, the effect of shear stress is less prominent than that in growth media, therefore resulting in correlation coefficient close to zero. In addition, a perfect positive correlation 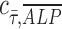 = 1 is represented by the dashed black line of slope 1 in Figure 2D-iii and iv. Points are scattered further away from the
= 1 is represented by the dashed black line of slope 1 in Figure 2D-iii and iv. Points are scattered further away from the 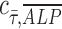 = 1 line in the osteogenic differentiation media condition than the points in the growth media condition, confirming greater positive correlation between ALP expression and shear stress under dynamic condition in growth media (Fig. 2D-iii) than in osteogenic media (Fig. 2D-iv).
= 1 line in the osteogenic differentiation media condition than the points in the growth media condition, confirming greater positive correlation between ALP expression and shear stress under dynamic condition in growth media (Fig. 2D-iii) than in osteogenic media (Fig. 2D-iv).
Flow can redistribute initially localized hMSCs near pillars
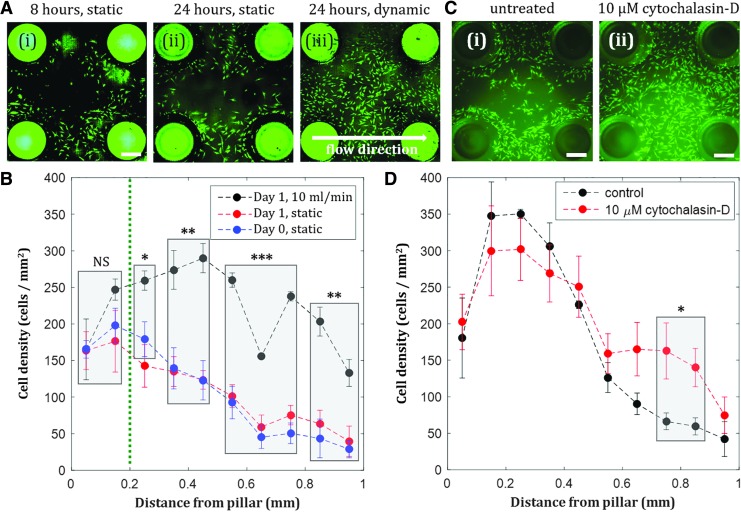
When hMSCs were seeded into the culture chambers, we observed initial cell localization around the pillars (Fig. 3A-i). After 24 h of static culture, hMSC localization was maintained (Fig. 3A-ii). Over time, cells proliferated and occupied the space further away from the pillars, with nearly uniform confluent coating of the bottom surface of the chamber observed by day 7 (Supplementary Fig. S2). Introduction of flow 4 h after cell seeding was observed to redistribute hMSCs that were initially localized near the pillars in the chambers, resulting in confluent coating of the bottom surface of the chamber after 24 h (Fig. 3A-iii). When the areal cell density inside the chamber was quantified at various distances from the pillars, we observed significant differences between the cell density of dynamic and static groups at distances >200 μm from the pillar (Fig. 3B). Therefore, the pillar-based culture chamber can be used to create localized hMSC populations, and the dynamic culture condition at early time points can optionally be applied depending on whether an isolated or uniform cell distribution is desired for subsequent experiments. While the use of perfusion bioreactors to enhance cell uniformity has been reported,6 our approach can utilize the coupling of flow and substrate curvature to switch the culture configuration from localized (Fig. 3A-i, ii) to uniform populations (Fig. 3A-iii). Our resulting spatial cell distribution was observed in a smaller, more physiologically relevant length scale than the cell distribution in other reported perfused constructs.46
FIG. 3.
Shear stress compensates initial actin-regulated hMSC localization near negative curvature substrates. (A) Fluorescent images of hMSCs inside culture chambers after 8 h of static condition (i), 24 h of static condition (ii), and 24 h of dynamic condition, flow rate = 10 mL/min (iii). Scale bar = 500 μm. (B) Effect of shear stress on hMSC spatial distribution in the chambers. The difference between static and dynamic growth are statistically significant for distances greater than 0.2 mm, compensating for initial localization of cells on the pillars. Day 0 static and day 1 static are not statistically different. (C) hMSCs inside culture chambers untreated (i) and with 10 μM cytochalasin-D (an actin inhibitor) treatment (ii) after 8 h of static condition. Scale bar = 500 μm. (D) More hMSCs are observed further away from the pillars when the cells are treated with 10 μM cytochalasin-D. Statistically significant difference in cell density was observed at distance >0.7 mm. Values are averages of four experiments and error bars are standard deviations (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001).
To further understand the mechanism behind cell localization near the pillars inside the chambers, we test the effect of a known actin inhibitor, cytochalasin-D, on the cell spatial distribution. We observed more cells distributed further away from the pillars when treated with cytochalasin-D (Fig. 3C, D), confirming that actin cytoskeletons regulate hMSC attachment and consequently localization around the pillars.
Pillar-based culture chambers can be utilized to create spatially patterned cocultures
Understanding interactions between two or more cell types is crucial due to the presence of multiple cell types in the bone marrow microenvironment. Using various cell–cell junctions and paracrine signaling, stem cells are able to communicate with niche cells, consequently regulating stem cell behavior.37,47 Specifically, attention has been devoted to studying the complex interaction between hMSCs and ECs, as they are two essential cell types present in the bone marrow microenvironment,47,48 and they form synergistic interactions in cocultures for vascularized bone tissue engineering applications.49–51 Studying interactions among ECs, undifferentiated hMSCs, and hMSC-derived osteoblastic cells in cocultures will aid the development of a prevascularized engineered bone construct, ensuring sufficient nutrient and oxygen delivery to the bone once implanted in a defect site.
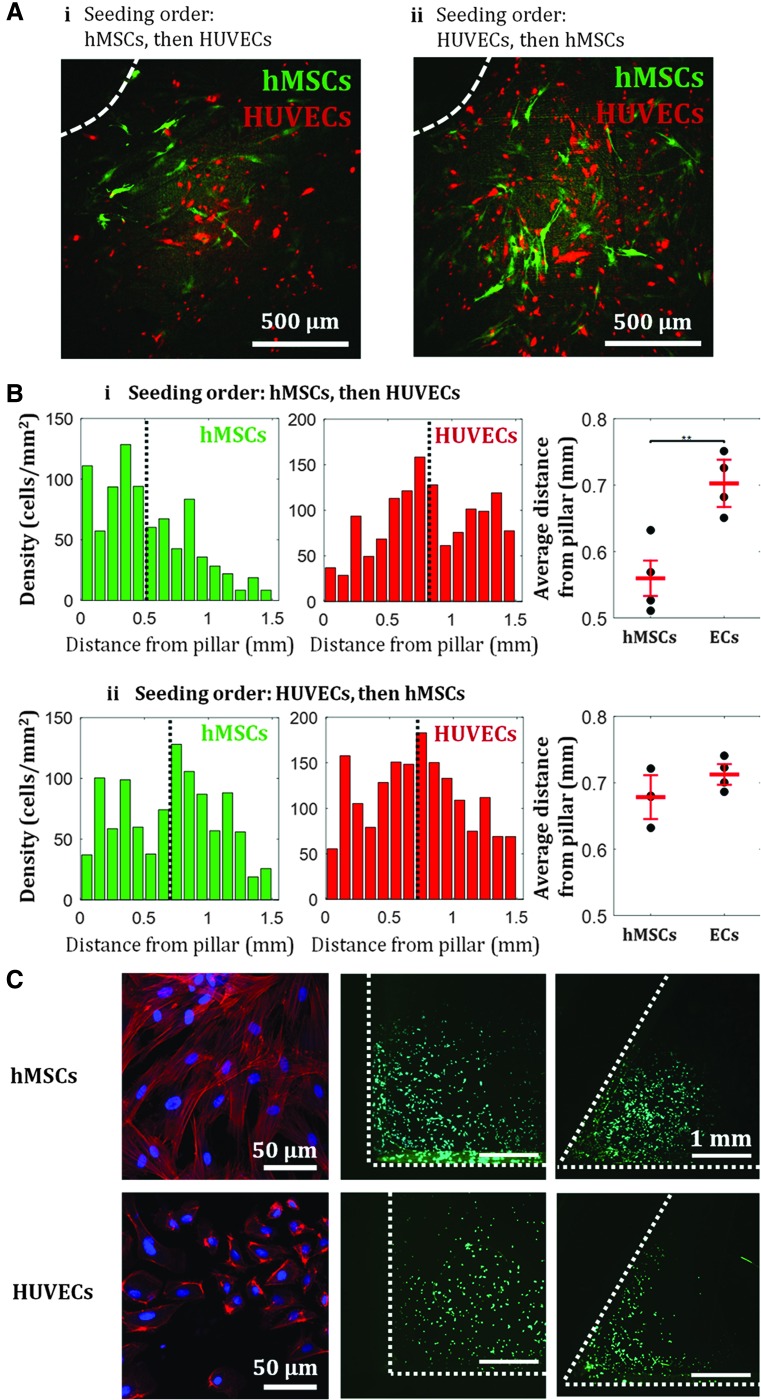
The observed initial crowding of hMSCs near the pillars resulted in spatially localized populations of hMSCs inside the chamber. We utilized this localization of hMSCs to create a spatially patterned coculture. Following hMSC seeding, cells were allowed to adhere for 12 h; HUVECs were then seeded afterward. As previously shown, hMSCs showed affinity toward the pillars, while HUVECs that were seeded later uniformly occupied the rest of the surface area between the pillars (Fig. 4A-i, B-i). Upon quantification of each cell type's density as a function of distance from the pillars, we found that hMSCs on average were located ∼550 μm from the pillar, while HUVECs were on average located ∼700 μm from the pillar (Fig. 4B-i). However, when the seeding order was reversed, we observed that both cell types were distributed more uniformly throughout the chamber surfaces (Fig. 4A-ii, B-ii). Using the same quantification, we found no statistical difference between the average distance from the pillar of hMSCs and of HUVECs (Fig. 4B-ii).
FIG. 4.
Spatial control of coculture of hMSCs and HUVECs in culture chambers. Seeding order determine the spatial distribution of the cells: hMSCs are localized near the pillars if seeded first (A-i), while both cell types are more uniformly distributed throughout the chamber if ECs are seeded first (A-ii). (A) Example of fluorescent images of hMSCs and HUVECs near pillars inside flow channel after 1 day of culture. Dashed white line indicates the edge of a pillar. (B) Cell density profile for when hMSCs are seeded first (i) and when HUVECs are seeded first (ii). Black dotted line indicates mean of the distribution. Means of the distributions indicate statistical difference between dominant growth/attachments regions of hMSCs and HUVECs when the hMSCs were seeded first, however, show no statistical difference when the HUVECs were seeded first. Values are averages of four experiments and error bars are standard deviations (**p ≤ 0.01). (C) Actin fibers configuration in hMSCs and HUVECs, and size-dependent cell attachment and localization. Red: F-actin, blue: nucleus, green: live cells. HUVECs localization is seen at corners with 60° angle, but not at 90° angle. EC, endothelial cell; HUVECs, human umbilical vein endothelial cells.
This result shows that the observed affinity toward negative curvature is only observed for hMSCs, not HUVECs. We hypothesize that the difference of attachment/localization behavior between the two cell types may be caused by the difference in cell size, which has been shown to correlate with F-actin content in various cell types.52,53 As hMSCs are larger than HUVECs, more prominent actin fibers are formed in hMSCs. While HUVECs showed no localization near the 90° angle between the pillars and the bottom surface, they localized in smaller angle corners (60°), indicating that cell preferential attachment and localization are size-dependent, where smaller cell size can sense smaller geometrical features (Fig. 4C, Supplementary Table S3). This result further confirms our earlier finding in Figure 3C that actin regulates hMSC attachment and localization near high curvature. With possible localization of one cell type around the pillar geometry through a specific seeding order, the culture chamber design can be used to tune hMSC-EC interactions by preferentially placing the pillars where hMSCs are desired.
Spatial configuration of cells in hMSC-EC cocultures dictates ATP-induced calcium signaling
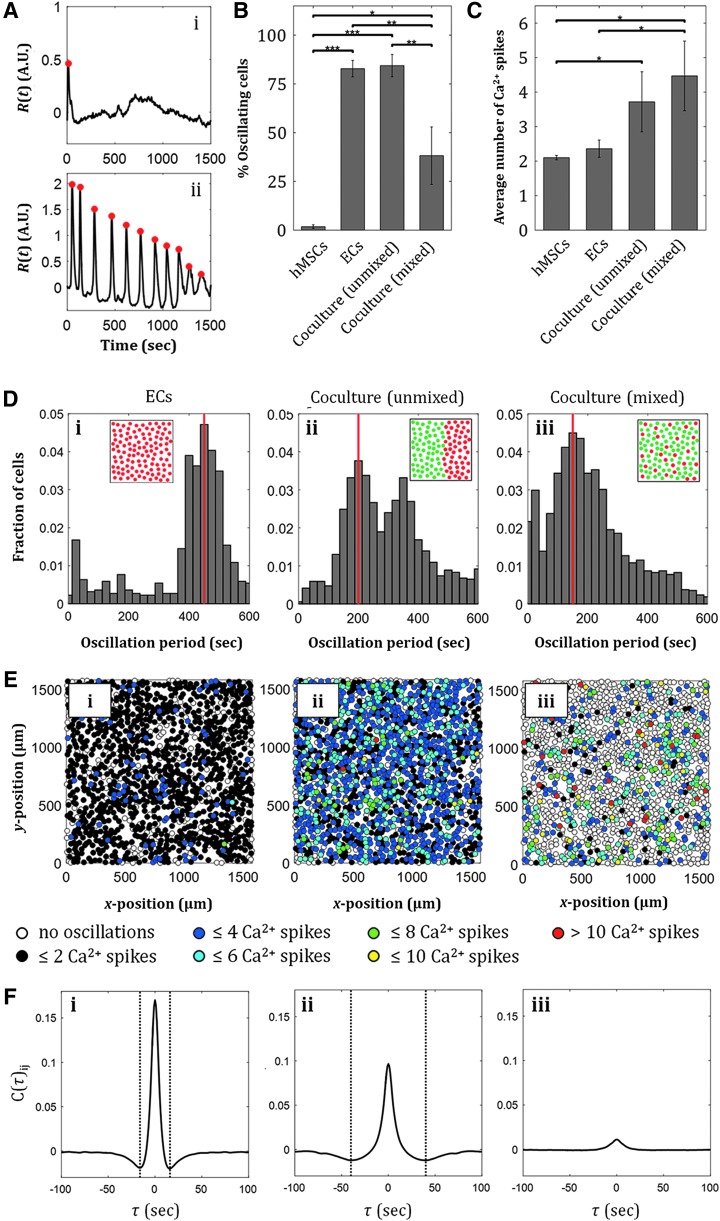
Through different localization behavior of hMSCs and ECs, signaling events between the two cell types can likely be modified by tuning the distance between the pillars or the pillar diameter, as the two have been shown to interact through distance-dependent paracrine signaling.50 We quantified calcium signaling in hMSC-EC cocultures in response to 10 μM ATP, an important common signaling molecule in various cellular processes, including proliferation and differentiation (Fig. 5 and Supplementary Fig. S3).54 Calcium signaling in hMSC and EC monocultures as well as in two configurations of cocultures (unmixed and mixed cocultures) were investigated. Upon ATP delivery to the cell cultures, we observed dynamic cellular Ca2+ response, as often shown by the oscillatory behavior of cytoplasmic Ca2+ level over time, R(t) (Fig. 5A). We quantified the fraction of cells that exhibited Ca2+ oscillations in each culture and observed that ECs in monoculture showed significantly more Ca2+ oscillations than hMSCs alone, while unmixed hMSC-EC coculture showed higher fraction of oscillating cells than hMSC-EC-mixed coculture (Fig. 5B and Supplementary Movies S1–S3). Out of those cells with oscillatory Ca2+ behavior, the two coculture configurations exhibited higher number of Ca2+ spikes than the monocultures (Fig. 5C). Quantification of oscillation period of each cell in each culture revealed distinct distribution of Ca2+ oscillation period for the EC monoculture and the two different spatial coculture configurations; EC monocultures showed oscillation period of 450 s, unmixed cocultures revealed two dominant oscillation periods at 200 and 350 s (Fig. 5D-ii and Supplementary Fig. S3A), while uniformly mixed cocultures showed only one dominant oscillation period (Fig. 5D-iii and Supplementary Fig. S3A). Note that this analysis is not relevant in hMSC monoculture due to the low number of cells that show Ca2+ oscillations. Although a higher fraction of cells in an unmixed coculture exhibited Ca2+ oscillations as shown by the number of colored cells (Fig. 5E-ii and Supplementary Fig. S3B) compared to cells in a mixed coculture, individual cells in a mixed coculture showed more Ca2+ spikes in 25 min during the response to ATP (Fig. 5E-iii) compared with individual cells in an unmixed coculture. The different spatial configurations of cocultures achieved in our chambers could therefore result in different Ca2+ signaling events, a mechanism known to be relevant in hMSC proliferation55 and osteogenic differentiation, where rapid Ca2+ oscillations in hMSCs have been shown to correlate with high levels of osteogenic gene markers.56
FIG. 5.
ATP-induced Ca2+ signaling in hMSC-EC coculture is affected by the cells' spatial configuration. (A) Examples of a cell exhibiting nonoscillating Ca2+ response (i), and a cell exhibiting Ca2+ oscillations (ii) in response to 10 μM ATP. The percentage of cells exhibiting Ca2+ oscillations varies in the two monocultures and the two different coculture configurations, with the unmixed coculture exhibiting the highest percentage of oscillating cells. (C) The average number of Ca2+ spikes in individual cells varies in the two monocultures and the two different coculture configurations, with the highest number of spikes in the mixed cocultures. Values are averages of three experiments and error bars are standard deviations (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001). (D) Distribution of Ca2+ oscillation periods in an unmixed hMSC-EC coculture reveals two dominant oscillation period (ii), while an EC monoculture and a mixed hMSC-EC coculture show only one dominant oscillation period (i, iii). Red line indicates modes of distributions. (E) More cells exhibit Ca2+ oscillations in an EC monoculture (i) and an unmixed coculture (ii), however, more Ca2+ spikes per cell are observed in a mixed coculture (iii). (F) Cross-correlation analysis for Ca2+ response of cells in EC monoculture (i), unmixed coculture (ii), and mixed coculture (iii): cellular communication is correlated with a time scale of 16 s for ECs, and 40 s for unmixed EC-MSC coculture, while Ca2+ responses in mixed populations show no temporal cross-correlation.
Furthermore, to understand cellular Ca2+ interactions within cultures upon response to ATP, we calculated the pair cross-correlation function C(τ)ij of the rate of change of calcium intensity, Ṙ(t), between all pairs of nearest neighbor cells (i, j) in a culture, where τ is the time delay between two Ca2+ responses of the corresponding neighboring cells.39,40 This density-dependent cross-correlation quantity has been quantified in dense cell populations that exhibit Ca2+ communication through gap junctions, where negative correlations were likely caused by locally transmitted signals from Ca2+ and second messengers between adjacent cells.39,40 In our experiments, EC monocultures, unmixed MSC-EC coculture, and mixed MSC-EC coculture reveal synchronization events as indicated by the positive correlation peak at τ = 0. However, the presence of a negative correlation peak, which has been shown to be indicative of gap-junctional communication timescale in cell populations,40 is only observed in the EC monoculture and the unmixed coculture (Fig. 5F). This negative correlation peak is observed at 16 s for EC monoculture and 40 s for unmixed hMSC-EC coculture.
Stem cell environment is reinforced by 3D cell culture formation and coculture of hMSCs and nonadherent blood-derived cell line
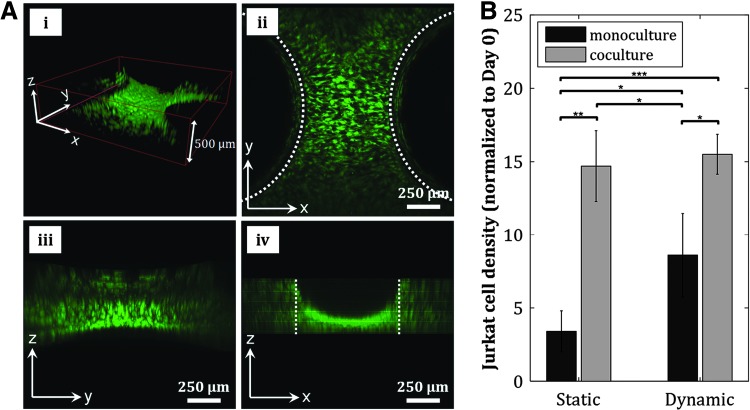
Formation of 3D cell cultures is physiologically relevant for tissue engineering applications. The pillar constructs inside our fluidic chamber enables the formation of 3D hMSC culture, as shown by the formation of 3D cell “bridges” structure between the pillars when the pillar-to-pillar distance was set to 500 μm (Fig. 6A and Supplementary Fig. S6B–D), rather than 2D cultures typically achieved in fluidic devices. The creation of a relevant model of the stem cell microenvironment in this culture chamber was also reinforced by the coculture of hMSCs and Jurkat cells, an immortalized T-lymphocyte cell line, inside this culture chamber. We seeded hMSCs in the chamber and perfused a suspension of Jurkat cells at a flow rate of 10 mL/min. Quantification of Jurkat cell density at static and dynamic conditions, and in monocultures and cocultures with hMSCs revealed enhancement of Jurkat cell proliferation when cocultured with hMSCs (Fig. 6B). Our fluidic chamber supports the culture of nonadherent blood-derived leukemia cells, suggesting the capability of using this dynamic culture system to study the growth of other blood-derived cells such as primary leukemia or lymphoid cells, or hematopoietic stem/progenitor cells (HSPCs) in this simple marrow organoid model. The Jurkat-hMSC synergistic interaction, along with 3D tissue formation, reinforces this culture chamber's capability of creating a relevant physiologic model of the stem cell microenvironment.
FIG. 6.
In vitro model of stem cell microenvironment is reinforced by 3D cell culture formation and coculture of hMSCs and Jurkat cells. (A) A 3D hMSC configuration is achieved by fabricating pillar constructs that are 500 μm apart from each other, as shown by the formation of cell “bridges” structure between the pillars in the 3D confocal reconstruction (i) and the corresponding confocal sections in x–y (ii), y–z (iii), and x–z (iv) planes. Dotted lines in (ii) and (iv) indicate edges of pillars. (B) Proliferation of Jurkat cells is enhanced in coculture with hMSCs (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001). 3D, three-dimensional.
Discussion
We have developed a 3D-printed fluidic culture chamber that serves as a platform for creating a 3D model stem cell microenvironment within a dynamic culture system for hMSCs. This single platform's capability to spatially pattern proliferation, differentiation, coculture, and form 3D cultures results from manipulation of pillar geometry and consequently flow pattern inside the chambers. We find that the proliferation of hMSCs in this platform under dynamic condition slightly exceeds the hMSC proliferation in two-dimensional static culture within 7 days. Our observation agrees with the reported enhancement of cell proliferation due to flow-induced shear stress on the cells, which is known to regulate hMSC proliferation through mitogen-activated protein kinase and calcium signaling.55 The range of shear stress experienced by the cells in our system is within the shear stress range, in which hMSC growth enhancement has been observed in 2D environments55,57 and 3D environments,7 with shear stress ranging from 0.1 to 1000 mPa, however, is closer to the lower end of the range reported in literature. This could explain the modest cell proliferative enhancement by dynamic culture in our system. Depending on the desired spatial distribution of cells for particular study applications, the introduction of flow at early time points during the culture can also be adjusted to give rise to either isolated cell populations of uniform monoculture.
Using flow-induced shear stress, our platform enables selective enhancement of osteogenic differentiation. Other methods of selective differentiation of MSCs into various lineages in tissue-engineered constructs have been reported, such as the use of mechanoelectrical stimulation in piezoelectric scaffolds,58,59 electrically conducting polymers,60 and electroactive microenvironments61,62 to direct differentiation into osteoblasts or chondrocytes, utilizing the tissue's capability of converting mechanical loads or deformation into electrical output.63,64 While electromechanical systems are attractive due to greater promoted differentiation than mechanical effects alone,58,62 our platform provides simplicity of use, where spatial patterning of osteogenic differentiation can be done simply through geometrically patterned flow, while simultaneously enhancing proliferation. In addition, even though tissues display electrical behavior due to the piezoelectric activities of extracellular matrix materials, most electromechanical approaches utilize piezoelectric materials as supporting 3D scaffolds to encapsulate cells, which then gets incorporated into a perfusion bioreactor, adding an additional step into the fabrication process of the tissue-engineered construct, while our approach seeds cells directly into the bioreactor platform whose geometry supports formation of 3D tissues.
In principle, future experiments can incorporate electromechanical stimuli through piezoelectric and/or elastic materials as the culture platform itself or as scaffolds to be inserted into our 3D-printed perfusion culture to further enhance patterned differentiation of specific cells. For example, human adipose stem cells cultured on a piezoelectric substrate, b-poly(vinylidene fluoride) shows more pronounced osteogenic differentiation when cultured on a “poled” surface (the negatively charged side of the material), with even higher expression of the osteogenic marker, ALP, when cultured dynamically in a bioreactor, where mechanical stimulation was applied through vibration at a frequency of 1 Hz.65 Mechanical stimuli other than shear has also been reported to enhance hMSC differentiation into other lineages; hMSCs encapsulated in alginate hydrogels exposed to periodic compressive force in a bioreactor shows enhancement of chondrogenic marker expression compared to those that are exposed to shear force.66 On the contrary, uniaxial cyclic strain increases smooth muscle marker expression from hMSCs.67,68 whereas equibiaxial static strain causes increased hMSC proliferation and production of vascular endothelial growth factor.69 One can imagine a culture system where combination of electromechanical stimulations and dynamic culture within a bioreactor is applied to create localized populations of specific cell types through placement of various cell-encapsulating materials with different responses. For example, 3D piezoelectric scaffolds can be used in conjunction with alginate beads in a bioreactor dynamic culture, where the piezoelectric scaffolds encapsulating hMSCs are localized in one area of the bioreactor and the hMSC-encapsulating alginate beads are in another. When applied during perfusion culture, compressive forces experienced by the alginate-encapsulated hMSCs during dynamic culture could enhance chondrogenic differentiation, while shear alone could enhance osteogenic differentiation of hMSCs embedded in the piezoelectric scaffold, or enhance hMSC/preosteoblastic cell proliferation.70,71 Such application provides one method to create two cell populations in a bioreactor chamber, such as biphasic osteochondral constructs. Applying such stimulations inside a fluidic bioreactor platform allows imaging to quantify the interaction between the two tissue types in a controlled environment. Introduction of various electromechanical stimuli into dynamic culture, combined with patterning capabilities facilitated by 3DP, provides innovative, custom approaches for the design of various tissue-engineered constructs.
Enhancement of ALP expression due to shear stress in our culture system is consistent with the increased expression of various osteogenic markers in other two-dimensional hMSC cultures under simple shear flow, with shear stresses ranging from 1 to 1000 mPa.72–76 The effect of shear stress on enhancement of ALP expression in hMSCs is more dominantly observed when the cells are cultured in growth media than in osteogenic media, as quantified by the positive correlation coefficient between ALP intensity and shear stress in growth media. Spatial patterning of hMSC differentiation at similar length scales have been done through mechanical stress gradient by creating multicellular “islands,” where regions of high stress near tissue edges resulted in osteogenesis, and regions of low stress away from edges resulted in adipogenesis.77 Our experiments support previous findings where osteogenic differentiation of MSCs is mediated by mechanosensitive molecules,78 as also shown in patterned MSC culture where regions of acute corners promoted osteogenesis due to increased actomyosin contractility.21 As an essential part of morphogenesis and tissue regeneration, MSCs undergo location-specific differentiation via cell–cell interactions mediated by direct contacts,79,80 making spatial positioning of the two cell type an important factor during differentiation. Our fluidic platform is therefore utilized to mimic the site-specific differentiation process, and the resulting patterned MSCs and osteoblasts within our device can be used to study the interactions between the two cell types.
When hMSCs are cultured dynamically in osteogenic media, ALP expression is observed throughout all surfaces of the culture chambers, regardless of the shear stress variation in those regions. The spatially homogeneous expression of ALP could be caused by the early presence of osteoblasts in the chamber during the culture due to the osteogenic media, and flow-induced shear stress further stimulates the osteoblast proliferation.81,82 These signaling events could explain the ALP expression in regions with almost zero shear stress in osteogenic media, resulting in nearly zero correlation between ALP expression and shear stress.
The preferential adherence of cells near the pillars under static conditions is consistent with the localization of cells near negative curvature substrates.83–85 Because cell–substrate is dictated by actin machinery and more prominent actin fibers have been observed on negatively curved substrates than on flat substrates,22 we perturbed this mechanism by inhibiting actin polymerization in hMSCs with cyctochalasin-D before seeding while maintaining viability (Supplementary Figs. S4 and S5 and Supplementary Tables S1 and S2), and quantified the cell localization. The assembly of filamentous stress fibers is known to be triggered by internal and/or external forces,86,87 suggesting a mechanism where mechanical forces arise in cells mainly in regions of high curvature, consequently stimulating cell proliferation.83,86 Our results show the lack of localization when hMSCs were treated with cytochalasin-D, confirming the involvement of actin in hMSC attachment and localization near negative curvatures, which is present in the intersections of the pillar surfaces and the bottom surface. Due to this localization, subsequent seeding of another cell type, in our case HUVECs, results in the two populations separated by ∼200 μm distance.
In addition to being responsive to curvature, MSCs also exhibit different commitment and differentiation signaling when cultured on substrates with different roughness, where osteogenic differentiation is favored on substrates with rough topography.88–90 Since 3DP results in submicron surface roughness,91,92 knowledge of the cells' mechanosensitivity to various substrate topographies could be utilized to spatially control osteogenic differentiation of MSCs in a bioreactor by 3DP composite scaffolds of two or more materials, which induce different cellular responses. This phenomenon, combined with our observation on curvature-driven cell localization, provides a simple method of creating spatially patterned cocultures of two cell types in our fluidic culture chamber for studies of cellular communications, such as calcium signaling as presented in our study, or other mechanosensitive cellular mechanisms.
The demonstrated ability to differentiate hMSCs into osteoblasts in particular locations via flow suggest that, in addition to providing enhanced cell numbers, we can use the pillar configuration in the culture chamber to tune the flow that eventually spatially patterns cell differentiation in tissues, which is vital to tissue regeneration. Moreover, we prove the capability to spatially pattern two cell types by exploiting one cell type's affinity toward the negative curvature surfaces near the pillars. Different strategies have been used to achieve cell patterning; in “macro” scale, populations of various cell types can be achieved by seeding strategies; however, this approach typically lacks control over cell–cell distance and thus the degree of cell–cell interaction. Gaining more control over cell–cell interaction at small, relevant scales, chemical and mechanical microenvironments have been successful through microfluidics. However, most PDMS-on-glass microfluidics were fabricated through complex, multistep lithography process to achieve specific geometries for the cell attachment, and most of the patterning can only be achieved in 2D.93 In our culture chamber, seeding strategies based on designed curved surfaces are used to establish patterned cell distributions, and subsequent application of flow further enables control over osteoblastic differentiation to generate more complex tissue environments. Localized osteoblast populations in the culture chamber can also be achieved through curvature-induced osteogenic differentiation,21 that is, by creating vertical geometrical structures that form acute angles with the bottom surface of the chamber to localize hMSCs around the structures upon initial seeding, and subsequently induce differentiation to osteoblasts due to the acute angle curvatures. Our patterning method can be used to tune heterotypic cell–cell interactions by simply utilizing one cell type's preferential adherence to curved substrates, while still maintaining the advantage of controlled microenvironments provided by fluidics.
One advantage of spatial patterning in our fluidic culture chamber is the ability to perform live cell mechanistic study of intercellular signaling, such as calcium signaling between hMSCs and ECs. The different spatial configurations of the two cell types result in various Ca2+ signaling behavior in response to ATP, as shown by the different Ca2+ spike numbers and oscillation periods between mixed and unmixed cocultures. Calcium oscillation behavior has been correlated to actin-mediated cellular activities,40,94 thus affecting actin-dependent processes in hMSCs,56,95 making it a crucial signaling event to quantify in our coculture. EC monocultures show both a high fraction of oscillating cells and a distinct negative peak in the cross-correlation analysis, indicating a prominent Ca2+ signaling among cells. In cocultures, such quantities are more prominent in spatially unmixed populations than in mixed cocultures, suggesting that cells need to be surrounded with other cells of the same type to generate a collective Ca2+ response. While hMSC monoculture shows low number of Ca2+ signaling (<2% of the monoculture show oscillatory Ca2+ behavior), hMSCs in unmixed cocultures become oscillatory in Ca2+ response to ATP, suggesting the beneficial effect of culturing them with ECs, particularly when spatially separated, to increase signaling. In addition, hMSC-EC cocultures show a shift in oscillation period from that of EC monocultures, suggesting capability of altering signaling dynamics through cell spatial positioning. This knowledge of spatial dependence of signaling could be applied to bone tissue engineering vascularization strategies, for example, by creating structures that mimic osteons in native cortical bones, where concentric bone-cell containing layers surround the central Haversian canal containing blood vessels.96 Since osteogenesis and vasculogenesis occur within this well-defined, separated cell population microenvironment, it is implicated that that cell positioning plays an important role in cell–cell interactions and regulation of cell phenotype. Further quantifications of Ca2+ signaling behavior and its downstream biological implications on hMSC and EC cellular processes are subjects of future studies. Other potential signaling mechanism in MSC-EC populations can be characterized by quantification of cytokines in media collected from the different monocultures and cocultures of ECs and MSCs.
Finally, the pillar constructs inside our fluidic chamber enable the formation of 3D hMSC culture, as shown by the cell growth on the pillars and the cell “bridges” structure between the pillars. The local growth of tissues around the local negative curvature caused by the pillars is consistent with previously reported local growth of various tissues in corners, followed by growth on the zero curvature surfaces as their neighborhood becomes curved.83,86 The platform's ability to form 3D tissues around the negatively curved substrates is an advantage in creating physiologically relevant 3D tissues from initial simple seeded monolayers, which is a major novelty of our platform. Many MSC culture systems have been developed for enhanced proliferation and osteogenic differentiation in 3D tissues; some recent developments include using nanoamplitude vibrations to generate mineralized matrix in collagen gels,97 modifying the perfusion duration of in vitro macroporous composite scaffold constructs containing MSCs to enhance their bone-forming capacity in vivo,98 using ceramic-encapsulated MSCs in perfusion bioreactor to reproducibly form bone in vivo,99 and coculturing MSCs and ECs in collagen scaffolds to enhance osteogenic differentiation.51 Our culture platform's geometry eliminates the need to fabricate porous supporting scaffolds that are required for 3D tissue configuration in these existing culture systems for MSC osteogenic differentiation. While detailed studies have been performed for bone tissue engineering applications where shear stress is evaluated throughout the constructs using computational fluid dynamic,100,101 the effect of stress on the cells is generally evaluated on a whole construct as an average,73,102 or on a much larger scale, for example, spatial variations in osteogenic differentiation within alginate scaffolds have been reported in the millimeter scale.19 Moreover, such studies present no clear correlation between this cellular effect and the mechanical environment. Our imaging capability in the fluidic culture platform, along with our analysis technique provides correlation between shear stress and cellular responses in the relevant length scales of ∼100 μm.
In reinforcing the culture chamber's capability to create a relevant model tissue microenvironment, we present a coculture between hMSCs and Jurkat cells, a T-lymphocyte cell line, showing enhancement of Jurkat cell proliferation in the presence of hMSCs, providing a simple model of coculture of MSCs and nonadherent blood cells in the bone marrow microenvironment. Cocultures of MSCs and other nonadherent blood cells or HSPCs are subjects of further studies in our culture chamber, as increased proliferation and reduced apoptosis of HSPCs have been reported when HSPCs are cultured with hMSCs.103,104
Moving forward, in scaling-up our fluidic culture chamber into a perfusion bioreactor system, an expanded bioreactor system incorporating multiple stacks of the pillar-based platform has been fabricated to provide three-dimensionality to cell culture with similar surface area to that of traditional culture plates. The expanded bioreactor construct maintains growth and viability of hMSCs in 3D structure after 7 days of dynamic culture (Supplementary Fig. S6). The supported growth of nonadherent blood-derived T cell leukemia cells in our culture chamber also encourages future study of growth, development, and physiology of hematopoietic cells in such 3D marrow organoid models, where a supportive stroma of MSC-EC coculture is present. Overall, by incorporating spatially patterned MSC-EC cocultures, flow-patterned osteogenic differentiation, culture of blood-derived cells, and the 3D tissue culture in our fluidic perfusion system, we are able to develop an in vitro tissue model incorporating features of the hematopoietic stem cell microenvironment in the bone marrow. Using tissue application-specific initial design in CAD, this fluidic culture platform can facilitate mechanistic studies of cell–cell and cell–substrate interactions through simple cell seeding strategies, without any material synthesis, scaffold assembly, or complex chemical modifications, and is scalable for larger stem cell expansion applications. Future applications of this system can include in vitro testing of the efficacy/toxicity of antineoplastic agents against (1) human cancer cells and (2) hematopoietic and stromal cells composing the hematopoietic niche.
Conclusions
We have developed a pillar-based fluidic perfusion culture chamber for dynamic hMSC culture that can be used for creating tissue model of stem cell environments, in addition to providing enhanced proliferation and osteogenic differentiation of hMSCs. The created in vitro microenvironment is achieved through spatially controlled proliferation and osteogenic differentiation of hMSCs, spatial patterning of hMSCs and ECs (two crucial components of the bone marrow niche) in coculture, and formation of 3D cultures. These are enabled via modulation of physiologic features of the native tissues such as geometry and fluid dynamic environment, which can be precisely tuned during our bioreactor fabrication through the ease of CAD and 3DP. While other fluidic platforms typically utilize supporting scaffolds for 3D cell culture, our system's associated geometry and flow patterns allow cell patterning and 3D cell culture simply from initial 2D seeding. Our study quantifies hMSC proliferation, osteogenic differentiation, their spatial dependence on local flow patterns and curvature in the culture chamber, along with spatial distribution achieved in an hMSC-EC coculture and the resulting modulation of calcium signaling, as well as demonstrates the 3D cell structure formation capability. The study highlights the culture chamber's potential as a simple innovative dynamic culture system for creating an in vitro model of a physiologic stem cell microenvironment, which can be easily modulated, scaled up and integrated into large-scale perfusion systems for relevant stem cell applications, or utilized to study various cell–cell and cell–substrate interactions. Such knowledge will be beneficial for future developments of bioreactors or scaffolds for tissue engineering and regenerative medicine applications.
Supplementary Material
Acknowledgments
The authors thank the University of Maryland Imaging Core Facility and Amy Beaven for the help with confocal microscopy. This work was supported by Maryland Stem Cell Research Fund/TEDCO under Award #2015-MSCRFI-1717 (to C.I.C. and J.P.F), by Maryland Industrial Partnerships grant (to J.P.F.), by Maryland Stem Cell Research Fund/TEDCO Postdoctoral Fellowship under Award #2017-MSCRFF-3920 (to J.L.), by NIST Fellowship program #2014-NIST-MSE-01 (to M.J.L.), and by the National Institute of Health under grant #P41 EB023833 (to J.P.F.).
Authors' Contributions
J.L., M.J.L., T.J.K., C.I.C., and J.P.F. designed research; J.L. and M.J.L. performed research; J.L., M.J.L., and J.P.F. analyzed data; and J.L., M.J.L., T.J.K., C.I.C., and J.P.F. wrote the article.
Disclosure Statement
T.J.K., C.I.C., and J.P.F. are founders of, and hold equity in, 3DBioWorks, Inc., which intends to develop bioreactors for cell culture. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of NIST.
References
- 1. Frith J.E., Thomson B., and Genever P.G. Dynamic three-dimensional culture methods enhance mesenchymal stem cell properties and increase therapeutic potential. Tissue Eng Part C Methods 16, 735, 2010. [DOI] [PubMed] [Google Scholar]
- 2. Occhetta P., Centola M., Tonnarelli B., Redaelli A., Martin I., and Rasponi M. High-throughput microfluidic platform for 3D cultures of mesenchymal stem cells, towards engineering developmental processes. Sci Rep 5, 10288, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Antoni D., Burckel H., Josset E., and Noel G. Three-dimensional cell culture: a breakthrough in vivo. Int J Mol Sci 16, 5517, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alvarez-Barreto J.F., Linehan S.M., Shambaugh R.L., and Sikavitsas V.I. Flow perfusion improves seeding of tissue engineering scaffolds with different architectures. Ann Biomed Eng 35, 429, 2007. [DOI] [PubMed] [Google Scholar]
- 5. Alvarez-Barreto J.F., and Sikavitsas V.I. Improved mesenchymal stem cell seeding on RGD-modified poly(l-lactic acid) scaffolds using flow perfusion. Macromol Biosci 7, 579, 2007. [DOI] [PubMed] [Google Scholar]
- 6. Wendt D., Marsano A., Jakob M., Heberer M., and Martin I. Oscillating perfusion of cell suspensions through three-dimensional scaffolds enhances cell seeding efficiency and uniformity. Biotechnol Bioeng 84, 205, 2003. [DOI] [PubMed] [Google Scholar]
- 7. Grayson W.L., Bhumiratana S., Cannizzaro C., Chao P.H., Lennon D.P., Caplan A.I., and Vunjak-Novakovic G. Effects of initial seeding density and fluid perfusion rate on formation of tissue-engineered bone. Tissue Eng Part A 14, 1809, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang J., Cao C., Wang W., Tong X., Shi D., Wu F., Zheng Q., Guo C., Pan Z., Gao C., and Wang J. Proliferation and osteogenesis of immortalized bone marrow-derived mesenchymal stem cells in porous polylactic glycolic acid scaffolds under perfusion culture. J Biomed Mater Res A 92, 817, 2010. [DOI] [PubMed] [Google Scholar]
- 9. Mygind T., Stiehler M., Baatrup A., Li H., Zou X., Flyvbjerg A., Kassem M., and Bünger C. Mesenchymal stem cell ingrowth and differentiation on coralline hydroxyapatite scaffolds. Biomaterials 28, 1036, 2007. [DOI] [PubMed] [Google Scholar]
- 10. Shin M., Yoshimoto H., and Vacanti J.P. In vivo bone tissue engineering using mesenchymal stem cells on a novel electrospun nanofibrous scaffold. Tissue Eng 10, 33, 2004. [DOI] [PubMed] [Google Scholar]
- 11. Sikavitsas V.I., Bancroft G.N., Lemoine J.J., Liebschner M.A., Dauner M., and Mikos A.G. Flow perfusion enhances the calcified matrix deposition of marrow stromal cells in biodegradable nonwoven fiber mesh scaffolds. Ann Biomed Eng 33, 63, 2005. [DOI] [PubMed] [Google Scholar]
- 12. Bancroft G.N., Sikavitsas V.I., van den Dolder J., Sheffield T.L., Ambrose C.G., Jansen J.A., and Mikos A.G. Fluid flow increases mineralized matrix deposition in 3D perfusion culture of marrow stromal osteoblasts in a dose-dependent manner. Proc Natl Acad Sci U S A 99, 12600, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Datta N., Pham Q.P., Sharma U., Sikavitsas V.I., Jansen J.A., and Mikos A.G. In vitro generated extracellular matrix and fluid shear stress synergistically enhance 3D osteoblastic differentiation. Proc Natl Acad Sci U S A 103, 2488, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grayson W.L., Fröhlich M., Yeager K., Bhumiratana S., Chan M.E., Cannizzaro C., Wan L.Q., Liu X.S., Guo X.E., and Vunjak-Novakovic G. Engineering anatomically shaped human bone grafts. Proc Natl Acad Sci U S A 107, 3299, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Janssen F.W., van Dijkhuizen-Radersma R., Van Oorschot A., Oostra J., de Bruijn J.D., and Van Blitterswijk C.A. Human tissue-engineered bone produced in clinically relevant amounts using a semi-automated perfusion bioreactor system: a preliminary study. J Tissue Eng Regen Med 4, 12, 2010. [DOI] [PubMed] [Google Scholar]
- 16. Sikavitsas V.I., Bancroft G.N., Holtorf H.L., Jansen J.A., and Mikos A.G. Mineralized matrix deposition by marrow stromal osteoblasts in 3D perfusion culture increases with increasing fluid shear forces. Proc Natl Acad Sci U S A 100, 14683, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wendt D., Stroebel S., Jakob M., John G.T., and Martin I. Uniform tissues engineered by seeding and culturing cells in 3D scaffolds under perfusion at defined oxygen tensions. Biorheology 43, 481, 2006. [PubMed] [Google Scholar]
- 18. Yeatts A.B., and Fisher J.P. Tubular perfusion system for the long-term dynamic culture of human mesenchymal stem cells. Tissue Eng Part C Methods 17, 337, 2011. [DOI] [PubMed] [Google Scholar]
- 19. Yeatts A.B., Geibel E.M., Fears F.F., and Fisher J.P. Human mesenchymal stem cell position within scaffolds influences cell fate during dynamic culture. Biotechnol Bioeng 109, 2381, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yu X., Botchwey E.A., Levine E.M., Pollack S.R., and Laurencin C.T. Bioreactor-based bone tissue engineering: the influence of dynamic flow on osteoblast phenotypic expression and matrix mineralization. Proc Natl Acad Sci U S A 101, 11203, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kilian K.A., Bugarija B., Lahn B.T., and Mrksich M. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc Natl Acad Sci U S A 107, 4872, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Werner M., Blanquer S.B., Haimi S.P., Korus G., Dunlop J.W., Duda G.N., Grijpma D.W., and Petersen A. Surface curvature differentially regulates stem cell migration and differentiation via altered attachment morphology and nuclear deformation. Adv Sci (Weinh) 4, 1600347, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pilia M., Guda T., Shiels S.M., and Appleford M.R. Influence of substrate curvature on osteoblast orientation and extracellular matrix deposition. J Biol Eng 7, 23, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sonam S., Sathe S.R., Yim E.K., Sheetz M.P., and Lim C.T. Cell contractility arising from topography and shear flow determines human mesenchymal stem cell fate. Sci Rep 6, 20415, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim S.H., Ahn K., and Park J.Y. Responses of human adipose-derived stem cells to interstitial level of extremely low shear flows regarding differentiation, morphology, and proliferation. Lab Chip 17, 2115, 2017. [DOI] [PubMed] [Google Scholar]
- 26. Dan P., Velot É., Decot V., and Menu P. The role of mechanical stimuli in the vascular differentiation of mesenchymal stem cells. J Cell Sci 128, 2415, 2015. [DOI] [PubMed] [Google Scholar]
- 27. El-Ali J., Sorger P.K., and Jensen K.F. Cells on chips. Nature 442, 403, 2006. [DOI] [PubMed] [Google Scholar]
- 28. Chan H.F., Zhang Y., Ho Y.P., Chiu Y.L., Jung Y., and Leong K.W. Rapid formation of multicellular spheroids in double-emulsion droplets with controllable microenvironment. Sci Rep 3, 3462, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chan H.F., Zhang Y., and Leong K.W. Efficient one-step production of microencapsulated hepatocyte spheroids with enhanced functions. Small 12, 2720, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Choi C.H., Wang H., Lee H., Kim J.H., Zhang L., Mao A., Mooney D.J., and Weitz D.A. One-step generation of cell-laden microgels using double emulsion drops with a sacrificial ultra-thin oil shell. Lab Chip 16, 1549, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Utech S., Prodanovic R., Mao A.S., Ostafe R., Mooney D.J., and Weitz D.A. Microfluidic generation of monodisperse, structurally homogeneous alginate microgels for cell encapsulation and 3D cell culture. Adv Healthc Mater 4, 1628, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vickerman V., Blundo J., Chung S., and Kamm R. Design, fabrication and implementation of a novel multi-parameter control microfluidic platform for three-dimensional cell culture and real-time imaging. Lab Chip 8, 1468, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu M.H., Huang S.B., Cui Z., Cui Z., and Lee G.B. A high throughput perfusion-based microbioreactor platform integrated with pneumatic micropumps for three-dimensional cell culture. Biomed Microdevices 10, 309, 2008. [DOI] [PubMed] [Google Scholar]
- 34. Gao Y.D., Majumdar D., Jovanovic B., Shaifer C., Lin P.C., Zijlstra A., Webb D.J., and Li D. A versatile valve-enabled microfluidic cell co-culture platform and demonstration of its applications to neurobiology and cancer biology. Biomed Microdevices 13, 539, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen S.Y.C., Hung P.J., and Lee P.J. Microfluidic array for three-dimensional perfusion culture of human mammary epithelial cells. Biomed Microdevices 13, 753, 2011. [DOI] [PubMed] [Google Scholar]
- 36. Huang S.B., Wu M.H., Wang S.S., and Lee G.B. Microfluidic cell culture chip with multiplexed medium delivery and efficient cell/scaffold loading mechanisms for high-throughput perfusion 3-dimensional cell culture-based assays. Biomed Microdevices 13, 415, 2011. [DOI] [PubMed] [Google Scholar]
- 37. Scadden D.T. The stem-cell niche as an entity of action. Nature 441, 1075, 2006. [DOI] [PubMed] [Google Scholar]
- 38. Calvi L.M., Adams G.B., Weibrecht K.W., Weber J.M., Olson D.P., Knight M.C., Martin R.P., Schipani E., Divieti P., Bringhurst F.R., Milner L.A., Kronenberg H.M., and Scadden D.T. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 425, 841, 2003. [DOI] [PubMed] [Google Scholar]
- 39. Sun B., Lembong J., Normand V., Rogers M., and Stone H.A. Spatial-temporal dynamics of collective chemosensing. Proc Natl Acad Sci U S A 109, 7753, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lembong J., Sabass B., Sun B., Rogers M.E., and Stone H.A. Mechanics regulates ATP-stimulated collective calcium response in fibroblast cells. J R Soc Interface 12, 20150140, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bird R.B., Stewart W.E., and Lightfoot E.N. Transport Phenomena, 2nd ed New York: John Wiley & Sons, Inc., 2007. [Google Scholar]
- 42. Dong J.D., Gu Y.Q., Li C.M., Wang C.R., Feng Z.G., Qiu R.X., Chen B., Li J.X., Zhang S.W., Wang Z.G., and Zhang J. Response of mesenchymal stem cells to shear stress in tissue-engineered vascular grafts. Acta Pharmacol Sin 30, 530, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hung B.P., Babalola O.M., and Bonassar L.J. Quantitative characterization of mesenchymal stem cell adhesion to the articular cartilage surface. J Biomed Mater Res A 101, 3592, 2013. [DOI] [PubMed] [Google Scholar]
- 44. McCoy R.J., and O'Brien F.J. Influence of shear stress in perfusion bioreactor cultures for the development of three-dimensional bone tissue constructs: a review. Tissue Eng Part B Rev 16, 587, 2010. [DOI] [PubMed] [Google Scholar]
- 45. Yeatts A.B., and Fisher J.P. Bone tissue engineering bioreactors: dynamic culture and the influence of shear stress. Bone 48, 171, 2011. [DOI] [PubMed] [Google Scholar]
- 46. Melchels F.P., Tonnarelli B., Olivares A.L., Martin I., Lacroix D., Feijen J., Wendt D.J., and Grijpma D.W. The influence of the scaffold design on the distribution of adhering cells after perfusion cell seeding. Biomaterials 32, 2878, 2011. [DOI] [PubMed] [Google Scholar]
- 47. Walker M.R., Patel K.K., and Stappenbeck T.S. The stem cell niche. J Pathol 217, 169, 2009. [DOI] [PubMed] [Google Scholar]
- 48. Yin T., and Li L. The stem cell niches in bone. J Clin Invest 116, 1195, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kaigler D., Krebsbach P.H., West E.R., Horger K., Huang Y.C., and Mooney D.J. Endothelial cell modulation of bone marrow stromal cell osteogenic potential. FASEB J 19, 665, 2005. [DOI] [PubMed] [Google Scholar]
- 50. Saleh F.A., Whyte M., Ashton P., and Genever P.G. Regulation of mesenchymal stem cell activity by endothelial cells. Stem Cells Dev 20, 391, 2011. [DOI] [PubMed] [Google Scholar]
- 51. Nguyen B.N.B., Moriarty R.A., Kamalitdinov T., Etheridge J.M., and Fisher J.P. Collagen hydrogel scaffold promotes mesenchymal stem cell and endothelial cell coculture for bone tissue engineering. J Biomed Mater Res A 105, 1123, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shankar J., and Nabi I.R. Actin cytoskeleton regulation of epithelial mesenchymal transition in metastatic cancer cells (vol 10, e0119954, 2015). PLoS One 10, e0119954, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sonowal H., Kumar A., Bhattacharyya J., Gogoi P.K., and Jaganathan B.G. Inhibition of actin polymerization decreases osteogeneic differentiation of mesenchymal stem cells through p38 MAPK pathway. J Biomed Sci 20, 71, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Berridge M.J., Lipp P., and Bootman M.D. The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol 1, 11, 2000. [DOI] [PubMed] [Google Scholar]
- 55. Riddle R.C., Taylor A.F., Genetos D.C., and Donahue H.J. MAP kinase and calcium signaling mediate fluid flow-induced human mesenchymal stem cell proliferation. Am J Physiol Cell Physiol 290, C776, 2006. [DOI] [PubMed] [Google Scholar]
- 56. Franz D., Karagaraj J., and Sun S. Matrix-mediated regulation of Ca2+ oscillations in osteodifferentiation of human mesenchymal stem cells. Phys Chem Stem Cells 1, 1, 2014. [Google Scholar]
- 57. Li Y.J., Batra N.N., You L., Meier S.C., Coe I.A., Yellowley C.E., and Jacobs C.R. Oscillatory fluid flow affects human marrow stromal cell proliferation and differentiation. J Orthop Res 22, 1283, 2004. [DOI] [PubMed] [Google Scholar]
- 58. Damaraju S.M., Shen Y., Elele E., Khusid B., Eshghinejad A., Li J., Jaffe M., and Arinzeh T.L. Three-dimensional piezoelectric fibrous scaffolds selectively promote mesenchymal stem cell differentiation. Biomaterials 149, 51, 2017. [DOI] [PubMed] [Google Scholar]
- 59. Sobreiro-Almeida R., Tamaño-Machiavello M.N., Carvalho E.O., Cordón L., Doria S., Senent L., Correia D.M., Ribeiro C., Lanceros-Méndez S., Sabater I, Serra R., Gomez Ribelles J.L., and Sempere A. Human mesenchymal stem cells growth and osteogenic differentiation on piezoelectric poly(vinylidene fluoride) microsphere substrates. Int J Mol Sci 18, E2391, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Guex A.G., Puetzer J.L., Armgarth A., Littmann E., Stavrinidou E., Giannelis E.P., Malliaras G.G., and Stevens M.M. Highly porous scaffolds of PEDOT:PSS for bone tissue engineering. Acta Biomater 62, 91, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li L., Yu M., Ma P.X., and Guo B. Electroactive degradable copolymers enhancing osteogenic differentiation from bone marrow derived mesenchymal stem cells. J Biomed Mater Res B 4, 471, 2016. [DOI] [PubMed] [Google Scholar]
- 62. Li Y., Dai X., Bai Y., Liu Y., Wang Y., Liu O., Yan F., Tang Z., Zhang X., and Deng X. Electroactive BaTiO3 nanoparticle-functionalized fibrous scaffolds enhance osteogenic differentiation of mesenchymal stem cells. Int J Nanomater 12, 4007, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Isaacson B.M., and Bloebaum R.D. Bone bioelectricity: what have we learned in the past 160 years? J Biomed Mater Res A 95, 1270, 2010. [DOI] [PubMed] [Google Scholar]
- 64. Fukada E., and Yasuda I. On the piezoelectric effect of bone. J Phys Soc Jpn 12, 1158, 1957. [Google Scholar]
- 65. Ribeiro C., Pärssinen J., Sencadas V., Correia V., Miettinen S., Hytönen V.P., and Lanceros-Méndez S. Dynamic piezoelectric stimulation enhances osteogenic differentiation of human adipose stem cells. J Biomed Mater Res A 103, 2172, 2015. [DOI] [PubMed] [Google Scholar]
- 66. Guo T., Yu L., Lim C.G., Goodley A.S., Xiao X., Placone J.K., Ferlin K.M., Nguyen B.N., Hsieh A.H., and Fisher J.P. Effect of dynamic culture and periodic compression on human mesenchymal stem cell proliferation and chondrogenesis. Ann Biomed Eng 44, 2103, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kurpinski K., Chu J., Hashi C., and Li S. Anisotropic mechanosensing by mesenchymal stem cells. Proc Natl Acad Sci U S A 103, 16095, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kurpinski K., Chu J., Wang D., and Li S. Proteomic profiling of mesenchymal stem cell responses to mechanical strain and TGF-beta1. Cell Mol Bioeng 2, 606, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kim I.S., Song Y.M., and Hwang S.J. Osteogenic responses of human mesenchymal stromal cells to static stretch. J Dent Res 89, 1129, 2010. [DOI] [PubMed] [Google Scholar]
- 70. Ribeiro C., Correia V., Martins P., Gama F.M., and Lanceros-Mendez S. Proving the suitability of magnetoelectric stimuli for tissue engineering applications. Colloids Surf B Biointerfaces 140, 430, 2016. [DOI] [PubMed] [Google Scholar]
- 71. Ribeiro C., Moreira S., Correia V., Sencadas V., Rocha J.G., Gama F.M., Gómez Ribelles J.L., and Lanceros-Méndez S. Enhanced proliferation of pre-osteoblastic cells by dynamic piezoelectric stimulation. RSC Adv 2, 11504, 2012. [Google Scholar]
- 72. Scaglione S., Wendt D., Miggino S., Papadimitropoulos A., Fato M., Quarto R., and Martin I. Effects of fluid flow and calcium phosphate coating on human bone marrow stromal cells cultured in a defined 2D model system. J Biomed Mater Res A 86, 411, 2008. [DOI] [PubMed] [Google Scholar]
- 73. Yourek G., McCormick S.M., Mao J.J., and Reilly G.C. Shear stress induces osteogenic differentiation of human mesenchymal stem cells. Regen Med 5, 713, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kreke M.R., Huckle W.R., and Goldstein A.S. Fluid flow stimulates expression of osteopontin and bone sialoprotein by bone marrow stromal cells in a temporally dependent manner. Bone 36, 1047, 2005. [DOI] [PubMed] [Google Scholar]
- 75. Delaine-Smith R.M., MacNeil S., and Reilly G.C. Matrix production and collagen structure are enhanced in two types of osteogenic progenitor cells by a simple fluid shear stress stimulus. Eur Cells Mater 24, 162, 2012. [DOI] [PubMed] [Google Scholar]
- 76. Grellier M., Bareille R., Bourget C., and Amédée J. Responsiveness of human bone marrow stromal cells to shear stress. J Tissue Eng Regen Med 3, 302, 2009. [DOI] [PubMed] [Google Scholar]
- 77. Ruiz S.A., and Chen C.S. Emergence of patterned stem cell differentiation within multicellular structures. Stem Cells 26, 2921, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lo Y.P., Liu Y.S., Rimando M.G., Ho J.H., Lin K.H., and Lee O.K. Three-dimensional spherical spatial boundary conditions differentially regulate osteogenic differentiation of mesenchymal stromal cells. Sci Rep 6, 21253, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Csaki C., Matis U., Mobasheri A., and Shakibaei M. Co-culture of canine mesenchymal stem cells with primary bone-derived osteoblasts promotes osteogenic differentiation. Histochem Cell Biol 131, 251, 2009. [DOI] [PubMed] [Google Scholar]
- 80. Kim H., Lee J.H., and Suh H. Interaction of mesenchymal stem cells and osteoblasts for in vitro osteogenesis. Yonsei Med J 44, 187, 2003. [DOI] [PubMed] [Google Scholar]
- 81. Kapur S., Baylink D.J., and Lau K.H. Fluid flow shear stress stimulates human osteoblast proliferation and differentiation through multiple interacting and competing signal transduction pathways. Bone 32, 241, 2003. [DOI] [PubMed] [Google Scholar]
- 82. Lee D.Y., Li Y.S., Chang S.F., Zhou J., Ho H.M., Chiu J.J., and Chien S. Oscillatory flow-induced proliferation of osteoblast-like cells is mediated by alpha(v)beta(3) and beta(1) integrins through synergistic interactions of focal adhesion kinase and Shc with phosphatidylinositol 3-kinase and the Akt/mTOR/p70S6K pathway. J Biol Chem 285, 30, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Rumpler M., Woesz A., Dunlop J.W., van Dongen J.T., and Fratzl P. The effect of geometry on three-dimensional tissue growth. J R Soc Interface 5, 1173, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lee Y.H., Huang J.R., Wang Y.K., and Lin K.H. Three-dimensional fibroblast morphology on compliant substrates of controlled negative curvature. Integr Biol (Camb) 5, 1447, 2013. [DOI] [PubMed] [Google Scholar]
- 85. Alias M.A., and Buenzli P.R. Modeling the effect of curvature on the collective behavior of cells growing new tissue. Biophys J 112, 193, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Nelson C.M., Jean R.P., Tan J.L., Liu W.F., Sniadecki N.J., Spector A.A., and Chen C.S. Emergent patterns of growth controlled by multicellular form and mechanics. Proc Natl Acad Sci U S A 102, 11594, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Putnam A.J., Cunningham J.J., Dennis R.G., Linderman J.J., and Mooney D.J. Microtubule assembly is regulated by externally applied strain in cultured smooth muscle cells. J Cell Sci 111, 3379, 1998. [DOI] [PubMed] [Google Scholar]
- 88. Balloni S., Calvi E.M., Damiani F., Bistoni G., Calvitti M., Locci P., Becchetti E., and Marinucci L. Effects of titanium surface roughness on mesenchymal stem cell commitment and differentiation signaling. Int J Oral Maxillofac Implants 24, 627, 2009. [PubMed] [Google Scholar]
- 89. Lee J.H., Shim W., Choolakadavil Khalid N., Kang W.S., Lee M., Kim H.S., Choi J., Lee G., and Kim J.H. Random networks of single-walled carbon nanotubes promote mesenchymal stem cell's proliferation and differentiation. ACS Appl Mater Interfaces 7, 1560, 2015. [DOI] [PubMed] [Google Scholar]
- 90. Dalby M.J., Gadegaard N., Tare R., Andar A., Riehle M.O., Herzyk P., Wilkinson C.D., and Oreffo R.O. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat Mater 6, 997, 2007. [DOI] [PubMed] [Google Scholar]
- 91. Waheed S., Cabot J.M., Macdonald N.P., Lewis T., Guijt R.M., Paull B., and Breadmore M.C. 3D printed microfluidic devices: enablers and barriers. Lab Chip 16, 1993, 2016. [DOI] [PubMed] [Google Scholar]
- 92. Walczak R., and Adamski K. Inkjet 3D printing of microfluidic structures—on the selection of the printer towards printing your own microfluidic chips. J Micromechanics Microeng 25, 1362, 2015. [Google Scholar]
- 93. Zervantonakis I.K., Kothapalli C.R., Chung S., Sudo R., and Kamm R.D. Microfluidic devices for studying heterotypic cell-cell interactions and tissue specimen cultures under controlled microenvironments. Biomicrofluidics 5, 13406, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Lembong J., Sabass B., and Stone H.A. Calcium oscillations in wounded fibroblast monolayers are spatially regulated through substrate mechanics. Phys Biol 14, 045006, 2017. [DOI] [PubMed] [Google Scholar]
- 95. Kim T.J., Seong J., Ouyang M., Sun J., Lu S., Hong J.P., Wang N., and Wang Y. Substrate rigidity regulates Ca2+ oscillation via RhoA pathway in stem cells. J Cell Physiol 218, 285, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Rho J.Y., Kuhn-Spearing L., and Zioupos P. Mechanical properties and the hierarchical structure of bone. Med Eng Phys 20, 92, 1998. [DOI] [PubMed] [Google Scholar]
- 97. Tsimbouri P.M., Childs P.G., Pemberton G.D., Yang J., Jayawarna V., Orapiriyakul W., Burgess K., González-García C, Blackburn G., Thomas D., Vallejo-Giraldo C., Biggs M.J.P., Curtis A.S.G., Salmerón-Sánchez M., Reid S., and Dalby M.J. Stimulation of 3D osteogenesis by mesenchymal stem cells using a nanovibrational bioreactor. Nat Biomed Eng 1, 758, 2017. [DOI] [PubMed] [Google Scholar]
- 98. Mitra D., Whitehead J., Yasui O.W., and Leach J.K. Bioreactor culture duration of engineered constructs influences bone formation by mesenchymal stem cells. Biomaterials 146, 29, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hoch A.I., Duhr R., Di Maggio N., Mehrkens A., Jakob M., and Wendt D. Expansion of bone marrow mesenchymal stromal cells in perfused 3D ceramic scaffolds enhances in vivo bone formation. Biotechnol J 12, 2017. DOI: 10.1002/biot.201700071 [DOI] [PubMed] [Google Scholar]
- 100. Ball O., Nguyen B.B., Placone J.K., and Fisher J.P. 3D printed vascular networks enhance viability in high-volume perfusion bioreactor. Ann Biomed Eng 44, 3435, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Guyot Y., Luyten F.P., Schrooten J., Papantoniou I., and Geris L. A three-dimensional computational fluid dynamics model of shear stress distribution during neotissue growth in a perfusion bioreactor. Biotechnol Bioeng 112, 2591, 2015. [DOI] [PubMed] [Google Scholar]
- 102. Nishimura I., Hisanaga R., Sato T., Arano T., Nomoto S., Ikada Y., and Yoshinari M. Effect of osteogenic differentiation medium on proliferation and differentiation of human mesenchymal stem cells in three-dimensional culture with radial flow bioreactor. Regen Ther 2, 24, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Mehrasa R., Vaziri H., Oodi A., Khorshidfar M., Nikogoftar M., Golpour M., and Amirizadeh N. Mesenchymal stem cells as a feeder layer can prevent apoptosis of expanded hematopoietic stem cells derived from cord blood. Int J Mol Cell Med 3, 1, 2014. [PMC free article] [PubMed] [Google Scholar]
- 104. Jones D.L., and Wagers A.J. No place like home: anatomy and function of the stem cell niche. Nat Rev Mol Cell Biol 9, 11, 2008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.