Figure 4:
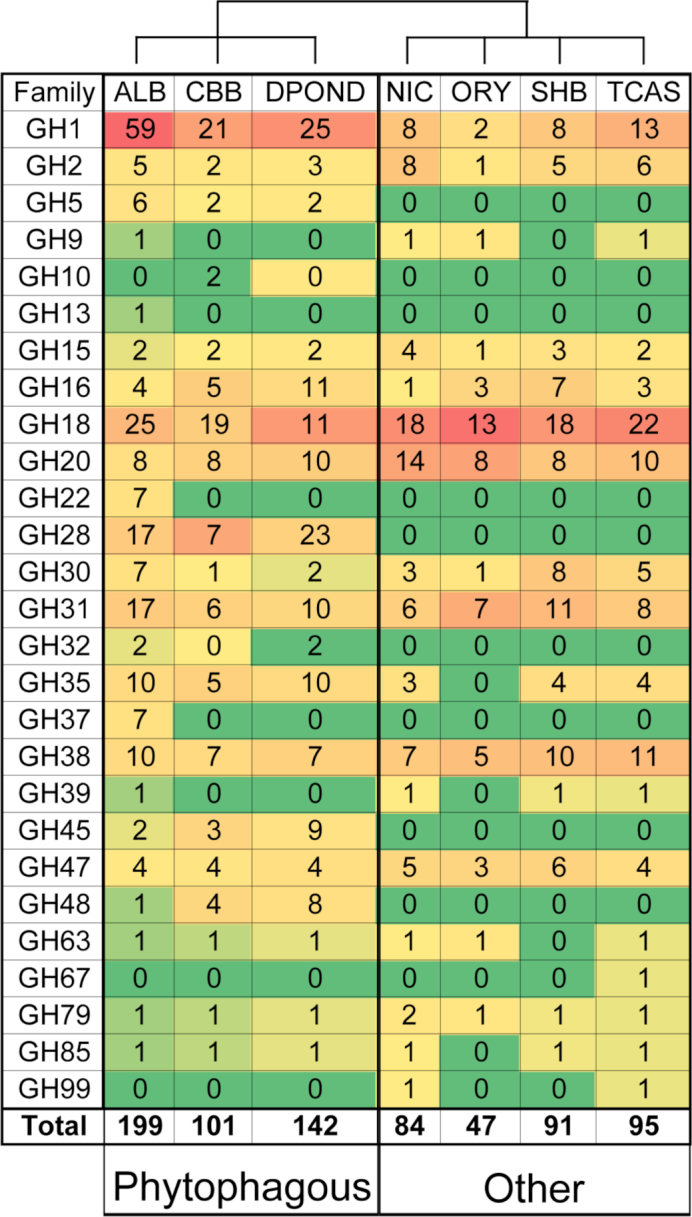
Glycoside hydrolase (GH) family copy numbers identified from beetle genomes. Genes coding for GHs were identified using Pfam domain assignments [24], and genome assemblies and coding gene predictions were obtained from NCBI (GenBank accession numbers: GCA_000390285.1 ALB, GCA_000355655.1 DPOND, GCA_001412225.1 NIC, GCA_001443705.1 ORY, GCA_000002335.3 TCAS) with the exception of CBB, which was downloaded from [25]. Families are color coded from green to red based on their relative abundance (total count/total number of GH genes), with red representing GH families that are highly abundant (≥25% of the total GH genes) and green representing GH families of lesser abundance (≤0.01%). Notably, the GH profiles of ATUMI and TCAST (neither of which feed on living plant material) differ strongly from the GH profiles of the phytophagous beetles, even though they all belong to the same infraorder, suggesting that diet, in part, might be driving the differences in GH family members and copy numbers. ALB = Asian longhorned beetle (A. glabripennis); CBB = Coffee berry borer (H. hampei); DPOND = Mountain pine beetle (D. ponderosae); NIC = burying beetle (N. vespilloides); ORY = scarab beetle (O. borbonicus); SHB = small hive beetle; and TCAS = red flour beetle (TCAST).
