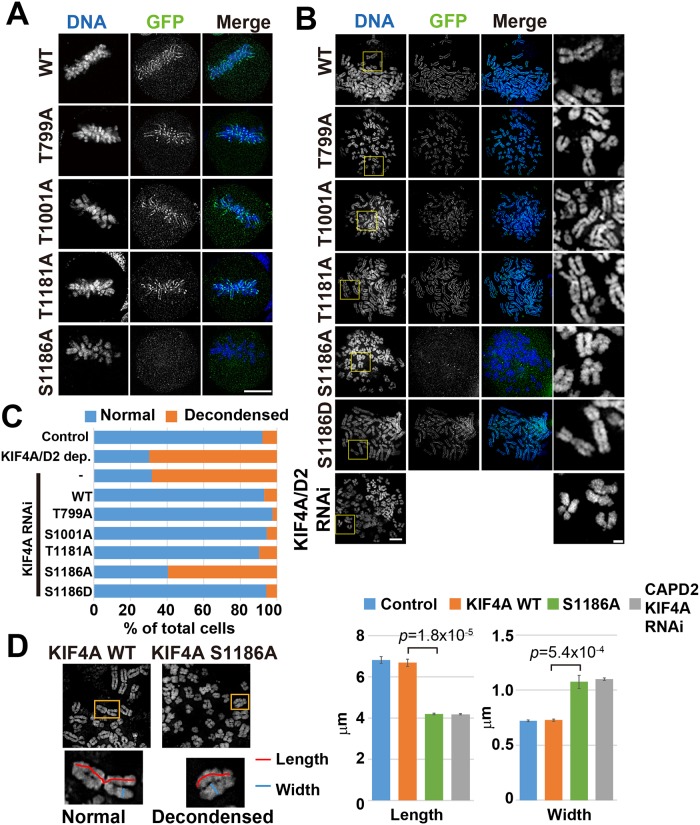
Fig 1. Phosphorylation of KIF4A S1186 is required for chromosome localization and chromosome morphology.
(A) Localization of EGFP-KIF4A WT or non-phosphorylatablemutants in metaphase cells. Only the KIF4A S1186 mutant did not localize to the mitotic chromosomes. Bar, 5 μm. (B) Localization of EGFP-KIF4A WT and mutants on spread chromosomes. Localization of KIF4A along the chromosome scaffold disappeared in EGFP-KIF4A S1186-expressing cells. Endogenous KIF4A was depleted in all EGFP-KIF4A-expressing cells. As a control for chromosome decondensation, spread chromosomes prepared from KIF4A and CAP-D2, a condensin I subunit, double depleted cells are indicated in the bottom panels. Images of stained DNA in yellow boxes are enlarged in the rightmost panels. Bars, 5 μm for whole images, 1 μm for enlarged images. (C) Percentage of cells showing normal and decondensed chromosome morphology, as shown in (D), in control (non-transfected), KIF4A/CAPD2 dep. (KIF4A and CAPD2 RNAi) (non-transfected), EGFP-KIF4A WT-, T799A-, S1001A-, T1181A-, S1186A-, and S1186D-expressing cells. More than 30 cells were counted for the classification in each condition, and the percentage reported is an average of three independent experiments. (D) Comparison of the chromosome size between EGFP-KIF4A WT- and S1186A-expressing cells. Entangled chromosomes are enlarged at the bottom of each image. The length (red) and width (blue) of the chromatid were measured. As a control for chromosome decondensation, chromosome size in both KIF4A and CAP-D2- depleted cells was also examined. The six longest chromosomes in a single cell were manually measured using ImageJ; 20 cells were analyzed in each condition. The graph shows the average length and width from three independent experiments.