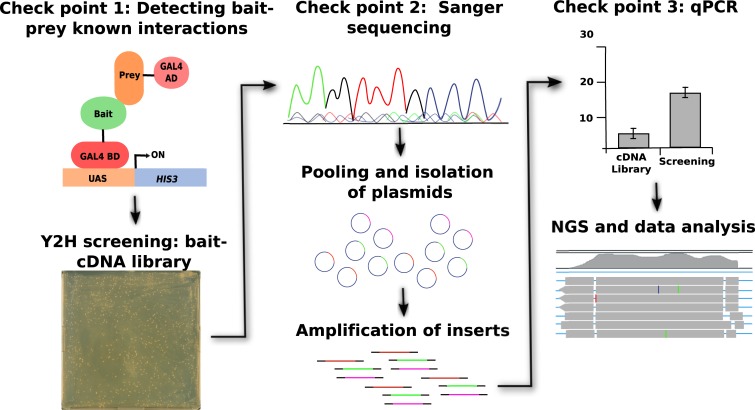
Fig 2. Y2H-seq workflow.
The first checkpoint involves analysis of bait functionality through a Y2H assay with known interactors (preys). Alternatively, immunoblot analysis can be carried out in checkpoint 1 to validate protein expression of the fusion proteins in case no interactors are known of a particular bait prior to the screen. Subsequently, the actual Y2H screening with the functional bait is carried out by supertransformation of the bait yeast strain with the prey cDNA library. In checkpoint 2, the plasmids are extracted and Sanger sequenced for some of the colonies obtained on the selective plates. Next, pooling of all colonies on the plates is carried out, all plasmids from the pool are isolated in a single extraction and the inserts of the plasmids are amplified by PCR. In checkpoint 3, qPCR is performed to verify for enrichment of expected preys with a particular bait Y2H-seq PCR mix. For that, semi-quantitative qPCR analysis is carried out relative to the PCR product of a screen with an empty bait vector or the cDNA library itself. Finally, NGS and data analysis is performed to obtain a final list of putative interactors of the interested bait proteins.