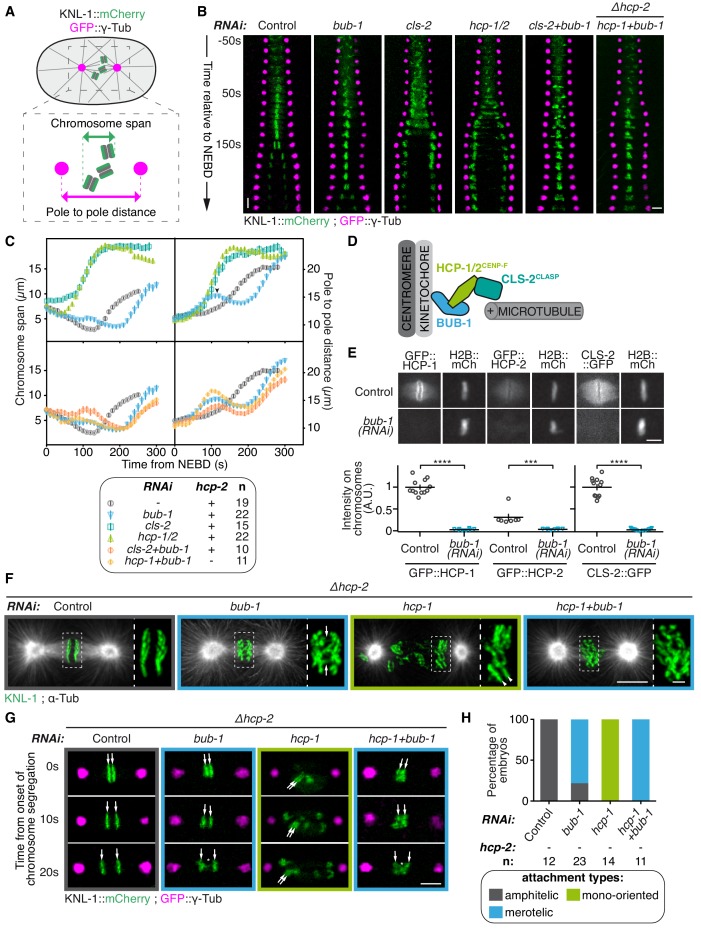
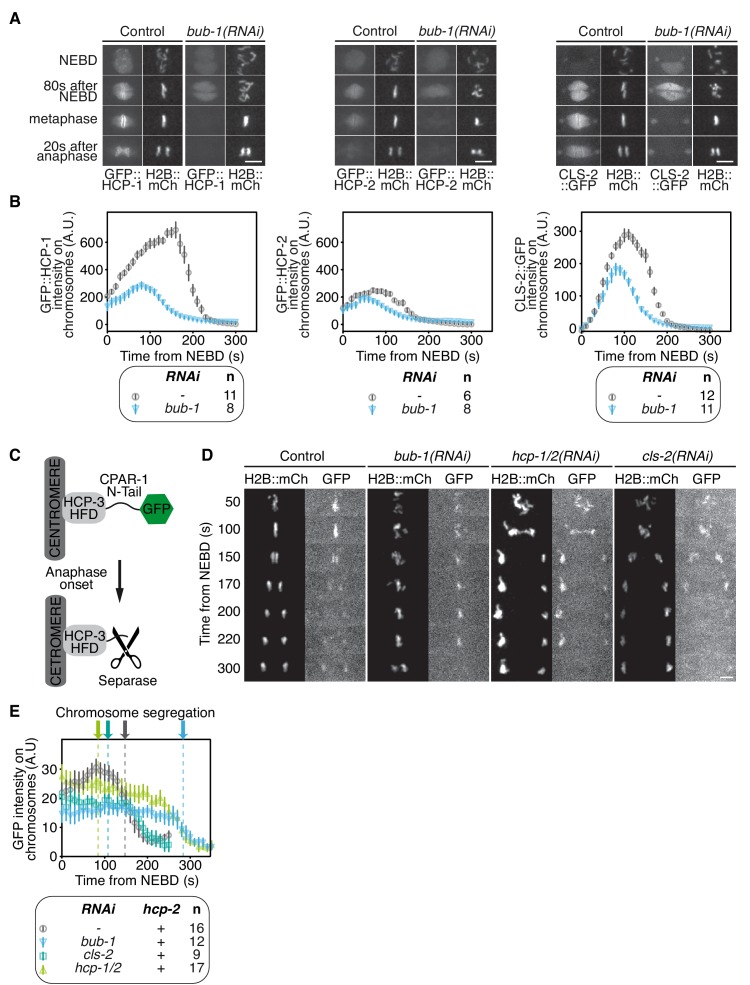
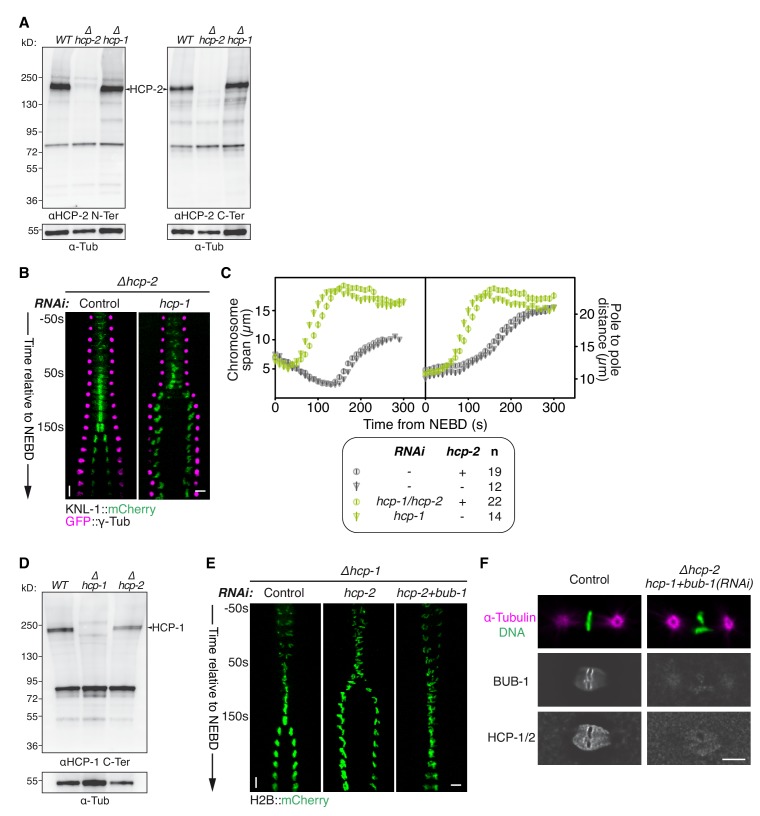
Figure 1. BUB-1 inhibits chromosome biorientation.
(A) Assay for kinetochore-microtubule attachment formation and chromosome congression. GFP::γ-Tub is used to measure the pole to pole distance, and KNL-1::mCherry is used to measure the chromosome span in the spindle pole axis. (B) Kymographs generated from embryos expressing GFP::γ-Tub and KNL-1::mCherry, for the different indicated conditions. Horizontal scale bar, 5 μm; Vertical scale bar, 20 s. (C) Chromosome span and pole to pole distance as functions of time after NEBD for the indicated conditions. Top right corner: Arrowhead, spindle pole bump following BUB-1 depletion. (D) Schematics of BUB-1 at kinetochores recruiting its downstream partners HCP-1/2CENP-F and CLS-2CLASP. (E) Top: Representative images from time-lapse movies showing BUB-1 dependent localisations of GFP::HCP-1CENP-F, GFP::HCP-2CENP-F and CLS-2CLASP::GFP on chromosomes (H2B::mCherry), at metaphase. Bottom: Quantification of the GFP signal on chromosomes at metaphase. Mann Whitney tests were used to determine significance (GFP::HCP-1 p < 0.0001, GFP::HCP-2 p = 0.0003, CLS-2::GFP p < 0.0001). Scale bar, 5 μm. (F) Immunofluorescent staining of kinetochores (KNL-1) and microtubules (DM1α) in Δhcp-2 zygotes at metaphase in the indicated conditions. Scale bar, 5 μm. Magnifications of the kinetochore region (highlighted by a dashed rectangle) are shown on the right of each panel. Arrows point to bent merotelic kinetochores in the BUB-1-depleted zygote. Arrowheads show a mono-oriented chromosome in the HCP-1CENP-F-depleted zygote. Scale bar, 1 μm. (G) Representative images of kinetochores (KNL-1::mCherry, green) and spindle poles (GFP::γ-Tub, magenta), at different times from the onset of chromosome segregation, for the indicated conditions. White arrows point towards sister kinetochores. White asterisks indicate the presence of kinetochore stretches. Scale bar, 5 μm. (H) Quantification of the percentage of embryos with chromosomes engaged in amphitelic, merotelic and mono-oriented attachments, in the indicated conditions. Error bars represent the SEM.