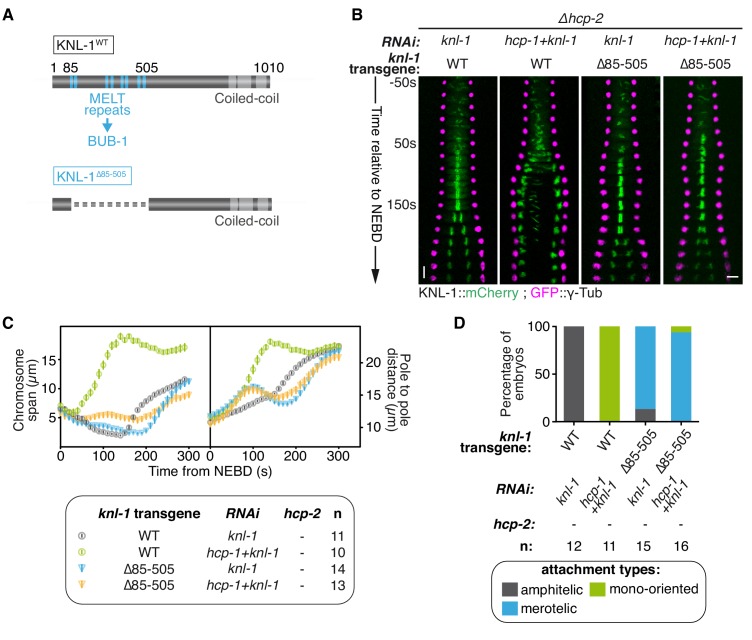
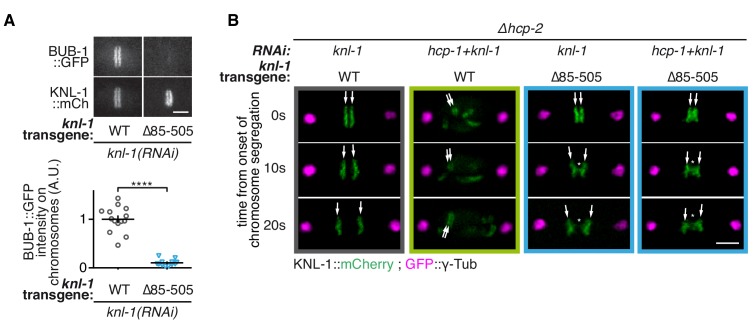
Figure 2. Biorientation inhibition requires BUB-1 localisation at the kinetochore.
(A) Schematics of WT KNL-1 and of the ∆85–505 mutant that leads to loss of BUB-1 from kinetochores. (B) Kymographs generated from embryos expressing GFP::γ-Tub and KNL-1::mCherry, for the indicated conditions. (C) Chromosome span and pole to pole distance as functions of time after NEBD for the indicated conditions. (D) Quantification of the percentage of embryos with chromosomes engaged in amphitelic, merotelic and mono-oriented attachments in the indicated conditions. Error bars represent the SEM. Horizontal scale bar, 5 μm; Vertical scale bar, 20 s.