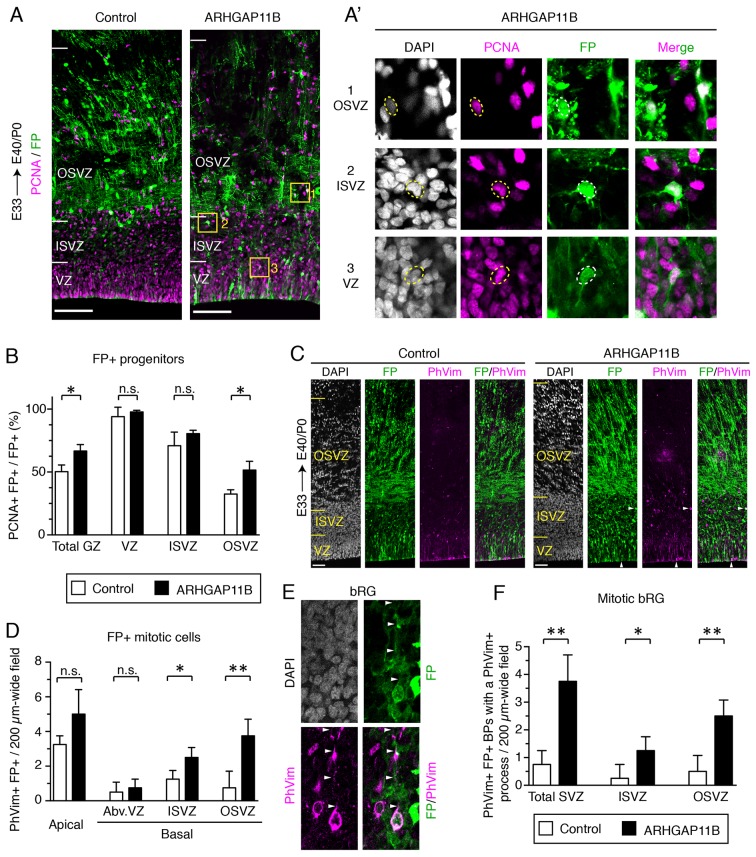
Figure 1. ARHGAP11B increases the abundance of BPs in the developing ferret neocortex.
Ferret E33 neocortex was electroporated in utero with a plasmid encoding a fluorescent protein (FP) together with either a plasmid encoding ARHGAP11B or empty vector (Control), followed by analysis at E40/P0. (A) Double immunofluorescence for FP (green) and PCNA (magenta) (for the images of the single channels and DAPI staining, see Figure 1—figure supplement 2A). Images are single optical sections. Scale bars, 100 μm. Boxes (50 × 50 μm) indicate FP+ BPs in the OSVZ (1, top), ISVZ (2, middle) and VZ (3, bottom), shown at higher magnification in (A). (A) Dashed lines indicate a cell body contour. (B) Percentage of FP+ cells in the germinal zones (GZ total) and in the VZ, ISVZ and OSVZ that are PCNA+ upon control (white) and ARHGAP11B (black) electroporations. Data are the mean of 3 experiments. Error bars indicate SD; *, p <0.05; n.s., not statistically significant; Student's t-test. (C) Double immunofluorescence for FP (green) and phospho-vimentin (PhVim, magenta), combined with DAPI staining (white). Images are single optical sections. Scale bars, 50 μm. Vertical arrowheads, apical mitosis; horizontal arrowheads, basal mitosis. (D) Quantification of FP+ mitotic cells, as revealed by PhVim immunofluorescence, in a 200 µm-wide field of the cortical wall, upon control (white) and ARHGAP11B (black) electroporations. Apical, mitoses lining the ventricular surface; basal, mitoses away from the ventricle (Abv.VZ, abventricular VZ; ISVZ; OSVZ). Data are the mean of 4 experiments. Error bars indicate SD; **, p <0.01; *, p <0.05; n.s., not statistically significant; Student's t-test. (E) Mitotic bRG (single optical sections). Double immunofluorescence for FP (green) and phospho-vimentin (PhVim, magenta), combined with DAPI staining (white), upon electroporation of the plasmid encoding FP together with the plasmid encoding ARHGAP11B. Arrowheads, PhVim+ basal process of the mitotic bRG. Images are oriented with the apical side facing down and are 25 μm wide. (F) Quantification of mitotic bRG (FP+ PhVim+ cell bodies in the SVZ that contain a PhVim+ process), in a 200 µm-wide field of total SVZ (left), ISVZ (middle) and OSVZ (right), upon control (white) and ARHGAP11B (black) electroporations. Data are the mean of 4 experiments. Error bars indicate SD; **, p <0.01; *, p <0.05; Student's t-test.