Main Text
Cells exploit various mechanisms to organize biochemical processes. Bringing together specific proteins and nucleic acids through condensation is now recognized as one of the key organizing mechanisms in eukaryotic cells (1, 2). Condensation of biomolecules results in dense, liquid-like bodies or domains in either the cytoplasm or the nucleoplasm, just like water vapor condenses to form cloud droplets (2). Inside these condensed bodies, specific proteins and nucleic acids can become highly concentrated, and they may experience a different chemical environment from the rest of the cell. Both effects can have profound effects on the availability, reactivity, and assembly behavior of individual proteins (3).
Recent years have seen a surge of examples of condensed liquid bodies in cells, commonly termed membraneless organelles (MLOs). They include nucleoli, paraspeckles, processing bodies, and stress granules (4). Most MLOs behave like liquid droplets and owe some of their unique properties to the fact that they lack a surrounding phospholipid membrane. They can fuse, flow, drip and dissolve, and take up matter from their surroundings freely. Nucleic acids are perhaps the single most important class of molecules that are taken up by MLOs. Increasing evidence suggests that nucleic acids (RNA and DNA) are by no means “inert” molecules inside MLOs but that their size, sequence, structure, and flexibility all matter. Nucleic acids have been found to affect MLOs in three different ways: they modulate the biophysical properties (5), induce selective nucleation (6), or recruit additional molecules (3). However, the respective roles of size, sequence, structure, and flexibility of the RNA or DNA in the modulation of MLOs remain unresolved.
To get a grip on this problem, systematic studies in model systems are crucial. In this issue, Shakya and King (7) shed light on the role of DNA flexibility in model MLOs by examining the appearance, stability, and dynamics of droplets of DNA and poly-L-lysine (PLL). Under suitable conditions, DNA and PLL, a disordered, cationic polypeptide, undergo liquid-liquid phase separation (LLPS), also known as complex coacervation. The phase separation is driven by attraction between opposite charges on DNA and PLL, whereas hydration and the disordered structure of the DNA and PLL chains ensure that the condensates remain liquid. The very same process of LLPS also underlies the formation of most MLOs (1, 3), although in those cases the driving forces are typically more diverse and often include cation-π and aromatic stacking interactions (2). Droplets of DNA and PLL have all the characteristics of liquid-condensed bodies in cells and are sensitive to salt. They provide an apt platform to investigate how the flexibility of nucleic acids affects LLPS.
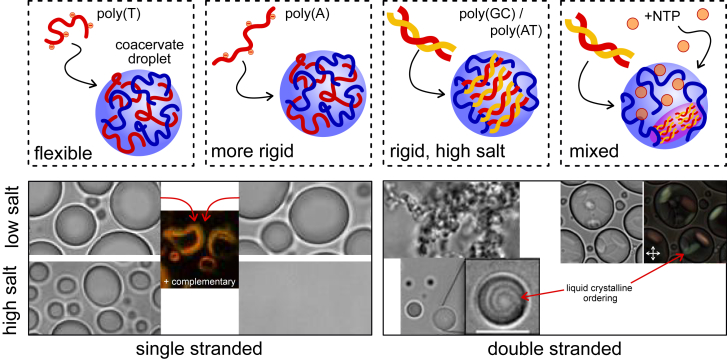
Shakya and King (7) first prepare droplets with single-stranded DNA (ssDNA) of 21 nucleotides and investigate if partial basepairing occurs inside the droplets. Adding complementary DNA strands leads to partial basepairing at the droplet edge, whereby a gel is locally formed, and the droplet is deformed (Fig. 1). Similar basepairing without droplet deformation has been observed for droplets of P-granule protein LAF-1 (5), but not for Ddx4 (8). In the latter case, existing basepairs were in fact disrupted, resulting in melting of short DNA duplexes inside the droplets. This difference likely stems from the aromatic residues in Ddx4 that are largely absent in both PLL and LAF-1. These residues can participate in favorable stacking interactions with free bases in unhybridized nucleic acids.
Figure 1.
Four experimentally studied cases of the role of nucleic acid flexibility in model MLOs. ssDNAs with different persistence lengths are visualized at low and high salt. Droplets with rigid chains dissolve more easily than droplets with flexible chains. dsDNA forms aggregates at low salt and liquid droplets with liquid-crystalline ordering at high salt. Mixing DNA with free NTPs leads to subcompartments that contain the liquid-crystalline phase. To see this figure in color, go online.
The authors subsequently focus on the role of ssDNA flexibility in these droplets. They compare the phase separation of poly(A) and poly(T) DNA strands of equal length and charge density but different flexibility. Poly(A), which has a larger persistence length than poly(T), forms weaker condensates that dissolve at lower salt concentration. How can the observed influence of DNA flexibility on LLPS be explained? This question may be answered by considering the charge neutralization that drives droplet formation by DNA and PLL. Polyelectrolytes, such as ssDNA, may need to bend to maximize their charge neutralization inside the droplets, which comes at an energy cost. For very rigid chains, the driving force may not be strong enough to bend them, leaving the chains in a conformation that does not achieve maximal charge neutralization. Both effects, bending and suboptimal charge neutralization, lead to reduced salt tolerance of the coacervate droplets. A simple analytical estimate of the first effect can be obtained by using a Debye-Hückel approximation. The average separation between opposite charges decreases upon condensation from κ−1 (the Debye screening length) to 1/d (the typical distance between ions in a pair), if counterion condensation is neglected. The corresponding energy gained in condensation can be written as (lB is the Bjerrum length, and α and β are constants) (9). Above a critical salt concentration ccrit, the energy gain is negligible, and condensed droplets dissolve, like Shakya and King (7) observe. The energy cost of bending a polymer with a curvature R is given by Ebend/kBT = πlp/R. It is higher for rigid chains with a larger persistence length lp. The energy gained in condensation is reduced by the cost of bending: ΔE′/kBT = (lB – εlp)/d – κlB, where ε = πd/R. This expression predicts that the critical salt concentration decreases with increasing persistence length as ccrit = β(1– εlp/lB)2, simply because it requires more energy to bend a more rigid chain. Droplets with poly(A) (lp ∼7.8 nm) are thus expected to have a 40% lower critical salt concentration (∼500 mM) compared to poly(T) (lp ∼3.1 nm), close to what Shakya and King (7) find experimentally.
By contrast, double-stranded DNA (dsDNA) does not form liquid droplets at low salt concentrations. Because dsDNA has a higher charge density, it binds much stronger to PLL and forms solid aggregates at low salt (Fig. 1). Upon increasing the salt concentration, liquid droplets are formed. More flexible poly(GC) (lp ∼41.7 nm) is able to form liquid droplets at significantly lower salt than more rigid poly(AT) (lp ∼50 nm) because it is more easy to bend to be accommodated in the liquid droplet phase. These findings are especially relevant in light of recent reports that LLPS may also mediate the formation of heterochromatin domains in the nucleus and thereby regulate gene expression (10). However, condensed heterochromatin is typically poor in GC, suggesting that the underlying condensation process is driven by more than just charge complexation. A striking feature of the PLL/dsDNA droplets in the work of Shakya and King (7) is that the dsDNA remains basepaired and forms a liquid-crystalline phase at high salt concentrations, as revealed by polarized light microscopy (Fig. 1). The dense dsDNA packing in liquid droplets bears some resemblance to DNA packing with similar liquid-crystalline ordering in virus capsids. Future experiments might explore these analogies further.
The work of Shakya and King (7) provides clear answers regarding the role of flexibility in liquid phase separation. These insights are valuable when interpreting observations on MLOs in vivo. However, it is inevitable that model systems simplify reality and disregard certain factors. In their work, Shakya and King only use short DNA, whereas MLOs contain mainly RNA, which can be much longer. Small but significant differences between the partitioning of RNA and DNA and a strong dependence on length have been reported (8). Does rigidity still rule for RNA droplets? More important perhaps are the role of additional types of interactions involved in MLOs and the flexibility of the condensed proteins. Does the flexibility of the nucleic acids still matter if the protein with which it interacts also contains structured domains like in nucleophosmin (4)? Cell biology still has many questions, but the power of biophysical experiments such as the work of Shakya and King is that they provide tools to look for the answers in a systematic way, one step at a time. Shakya and King have taken the first step forward.
Editor: Anatoly Kolomeisky.
References
- 1.Harmon T.S., Holehouse A.S., Pappu R.V. To mix, or to demix, that is the question. Biophys. J. 2017;112:565–567. doi: 10.1016/j.bpj.2016.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banani S.F., Lee H.O., Rosen M.K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gomes E., Shorter J. The molecular language of membraneless organelles. J. Biol. Chem. 2018:1–22. doi: 10.1074/jbc.TM118.001192. Published online July 25, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitrea D.M., Kriwacki R.W. Phase separation in biology; functional organization of a higher order. Cell Commun. Signal. 2016;14:1. doi: 10.1186/s12964-015-0125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elbaum-Garfinkle S., Kim Y., Brangwynne C.P. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. USA. 2015;112:7189–7194. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langdon E.M., Qiu Y., Gladfelter A.S. mRNA structure determines specificity of a polyQ-driven phase separation. Science. 2018;360:922–927. doi: 10.1126/science.aar7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shakya A., King J.T. DNA local flexibility dependent assembly of phase separated liquid droplets. Biophys. J. 2018;115:1840–1847. doi: 10.1016/j.bpj.2018.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nott T.J., Craggs T.D., Baldwin A.J. Membraneless organelles can melt nucleic acid duplexes and act as biomolecular filters. Nat. Chem. 2016;8:569–575. doi: 10.1038/nchem.2519. [DOI] [PubMed] [Google Scholar]
- 9.Spruijt E., Sprakel J., van der Gucht J. Relaxation dynamics at different time scales in electrostatic complexes: time-salt superposition. Phys. Rev. Lett. 2010;105:208301. doi: 10.1103/PhysRevLett.105.208301. [DOI] [PubMed] [Google Scholar]
- 10.Strom A.R., Emelyanov A.V., Karpen G.H. Phase separation drives heterochromatin domain formation. Nature. 2017;547:241–245. doi: 10.1038/nature22989. [DOI] [PMC free article] [PubMed] [Google Scholar]