Fig. 2.
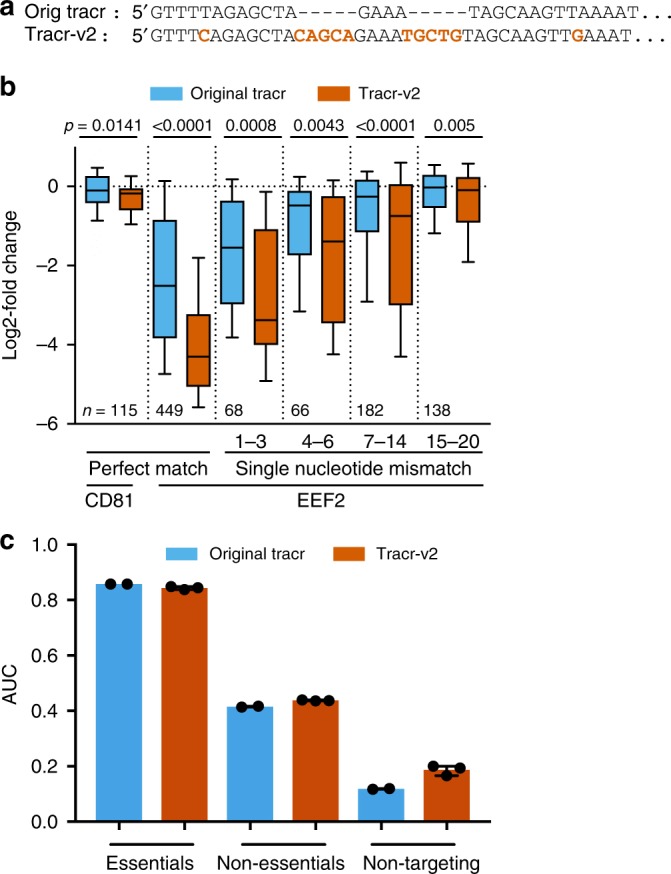
Evaluation of an alternative tracrRNA. a Comparison of the sequence of the original tracrRNA and tracr-v2. b Comparison of the log2-fold-change of perfectly matched sgRNAs and single base mismatches targeting the essential gene EEF2 for the original tracrRNA and tracr-v2. CD81-targeting sgRNAs serve as the control. Mismatched sgRNAs are binned by position, and are numbered such that nt 20 is PAM-proximal. The box represents the 25th, 50th, and 75th percentiles, and whiskers show 10th and 90th percentiles. Two-tailed Welch’s t-test was used to compare the distributions within each bin and p-values are indicated.The number of sgRNAs in each comparison is shown at the bottom. c AUC values for essential, non-essential, and non-targeting gene sets with the original tracrRNA and tracr-v2 in genome-wide viability screens. Error bars represent the range of two or three biological replicates for screens with the original and modified tracrRNA, respectively
