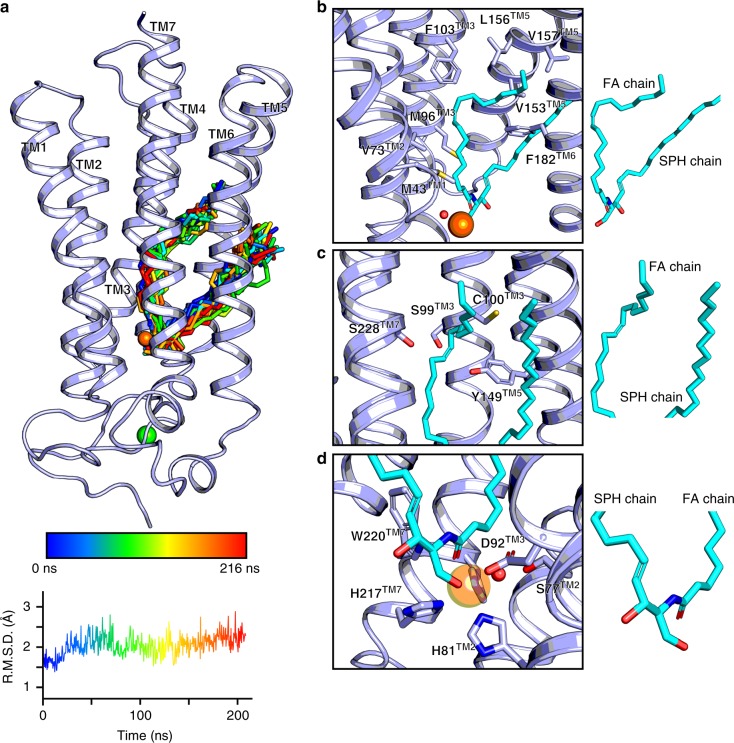
Fig. 3.
Computational analyses of ceramide 18:1 docking into ACER3. a Top scoring C18:1 ceramide (shown as stick) docking pose (hydrogens were omitted for clarity) and observed ligand trajectory during a 216 ns long MD simulations (from blue to red with ligand snapshots extracted every 8 ns). A representative all-atom RMSD of the C18:1 ceramide is shown below (n = 5 in total). b View of the ceramide docking pose highlighting as sticks the residues in close proximity to the ceramide (colored in cyan, and also shown alone on the right to indicate the identity of the fatty acid (FA) and sphingosine (SPH) chains). c Environment around the site of unsaturation of the docked C18:1 ceramide with the side chains of polar residues close to the sp2 carbons shown as sticks. d Close-up view of the Zn2+ catalytic site (side chains represented as sticks) with the putative water molecule (red sphere) ideally positioned to attack the amide bond of the C18:1 ceramide. In all panels, the Zn2+ and Ca2+ as well as the water molecule are represented as spheres (colored orange, green, and red, respectively)