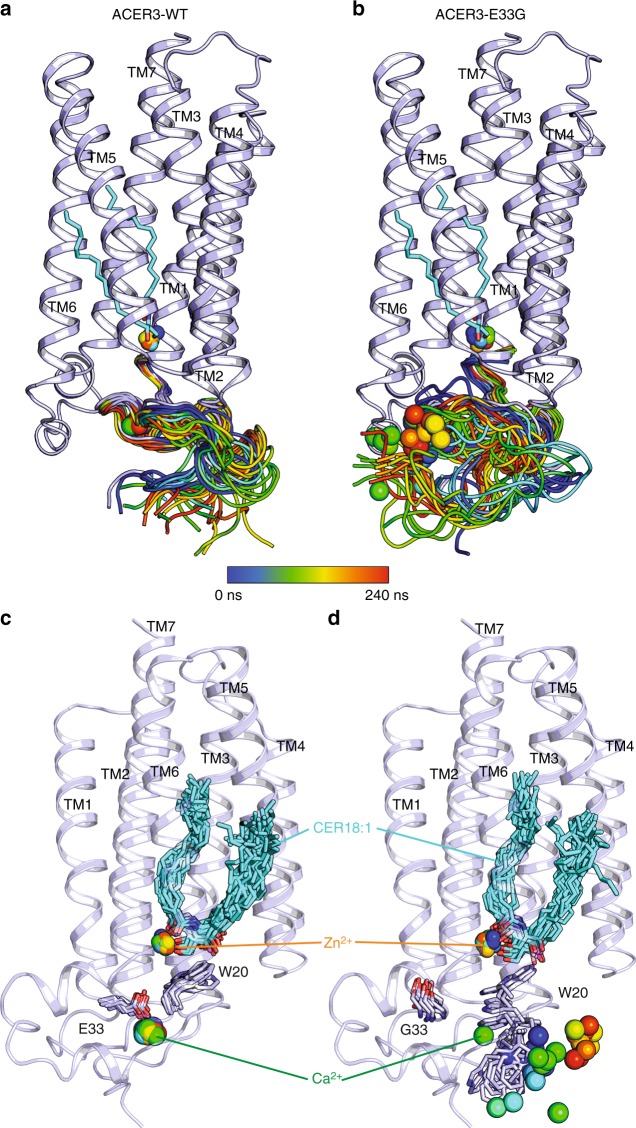
Fig. 6.
MD simulations of ACER3 wild-type and E33G mutant Snapshots of the calculated N-terminus domain trajectories during a 240 ns long MD simulation (from blue to red, taken every 10 ns) for the wild-type (a) or E33G mutant (b) showing that the mutation leads to a more dynamic/unstable N-terminus domain. c, d Snapshots of the ACER3 wt (c) or E33G mutant (d) showing the key ligands (Cer18:1, Zn2+ and Ca2+) and contacting residues (shown as sticks) trajectories during a 200 ns long MD simulations colored as above and highlighting the motion/instability of the W20 side chain and of the Ca2+ in the E33G mutant (d) in contrast to the wild-type ACER3 (c)