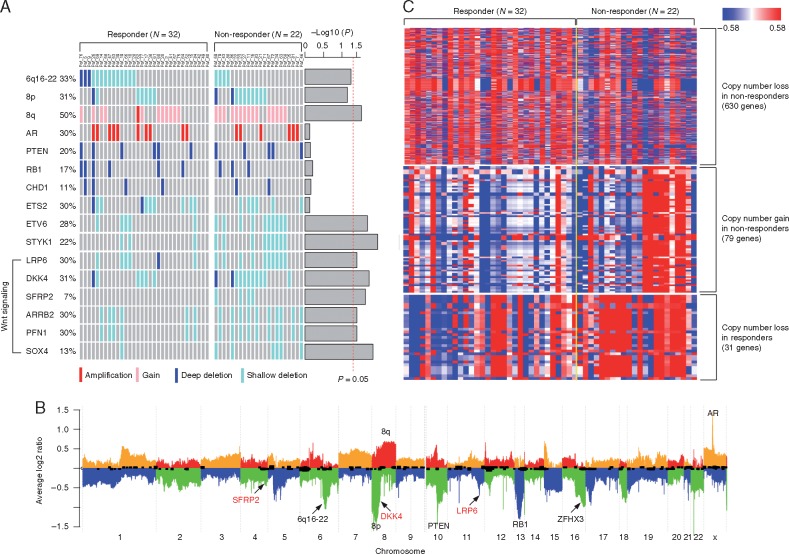
Figure 2.
(A) Comparing the arm-level (6q16-22, 8p, and 8q) and gene level copy number alteration frequencies between responders (N = 32) and nonresponders (N = 22). P-values were calculated using logistic regression and indicated as barplot on the right. The vertical dash line indicated P = 0.05. (B) Heat map showing genes with different copy number alterations (P < 0.05) between responders (N = 32) and nonresponders (N = 22). (C) Average copy number profiles of the 54 tumor genomes. Individual chromosomes are separated by alternating colors with ‘orange’ and ‘red’ indicating copy number gain and ‘blue’ and ‘green’ indicating copy number loss. Key aberrant genes and segments are indicated. Wnt pathway inhibitors (SFRP2, DKK4, and LRP6) are indicated in red.