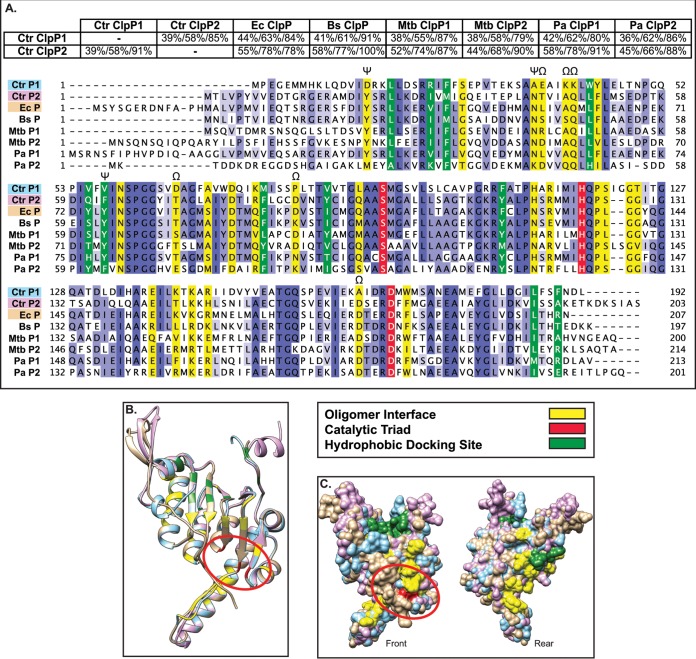
FIG 2.
Bioinformatics analyses suggest that both chlamydial ClpP paralogs are functional proteases (A). Pairwise alignments were performed using NCBI BLAST (default settings) and presented as percent identity/percent similarity/percent coverage. Multiple-sequence alignment was performed using Clustal Omega with default settings and presented using Jalview version 2. Organisms included are C. trachomatis (Ctr), E. coli (Ec), B. subtilis (Bs), M. tuberculosis (Mtb), and P. aeruginosa (Pa). Conserved residues are highlighted in various shades of blue depending on conservation strength across species, with the catalytic triad, oligomer interface, and hydrophobic docking region colored as indicated in the key. Residues in which ClpP1 has a radically different substitution compared to other ClpP proteins are denoted by Ω. Residues involved in activation conformational changes are denoted by ψ (33). (B and C) Predicted 3D structures acquired from Phyre2 were aligned and colored in Chimera (UCSF). (B) 3D structural alignment of C. trachomatis ClpP1, C. trachomatis ClpP2, and E. coli ClpP (peptide colors as shown in multiple-sequence alignment). Active-site residues are circled in red. (C) Space-filling model of predicted 3D structures. “Rear” is a 180° rotation around the y axis to show the surface of the protein not shown in the “Front” image.