Fig. 1.
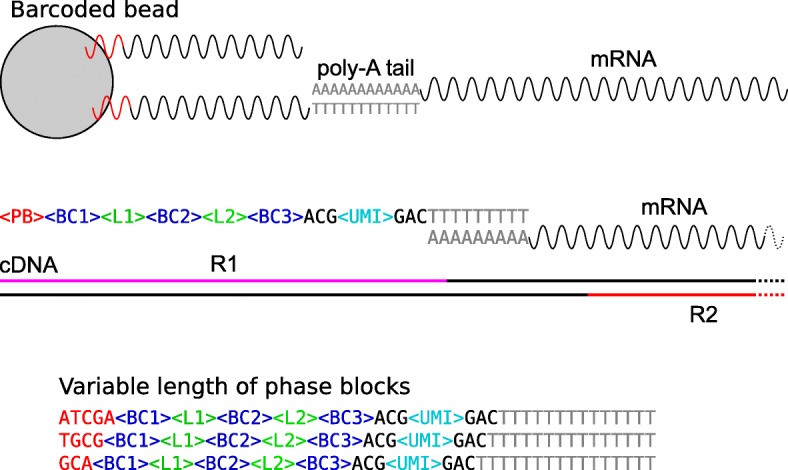
Schematic of Bio-Rad ddSEQ/Illumina reads’ structure (Top) In BioRad ddSEQ, barcoded beads capture mRNA molecules through hybridization with mRNA poly-A tails. Each single DNA strand is characterized by the following structure: a phase block (PB), three barcode blocks (BC1, BC2, BC3) interlinked by two different linkers (L1 and L2), and one UMI flanked by two trinucleotides (ACG and GAC). (Middle) Read 1 (R1) contains molecular tags while Read 2 (R2) contains the information of the mRNA sequence (R1 and R2 are not in scale). (Bottom) Separation of cDNA from the beads can occur at different nucleotides within the PB, thus making the position of the two linkers variable
