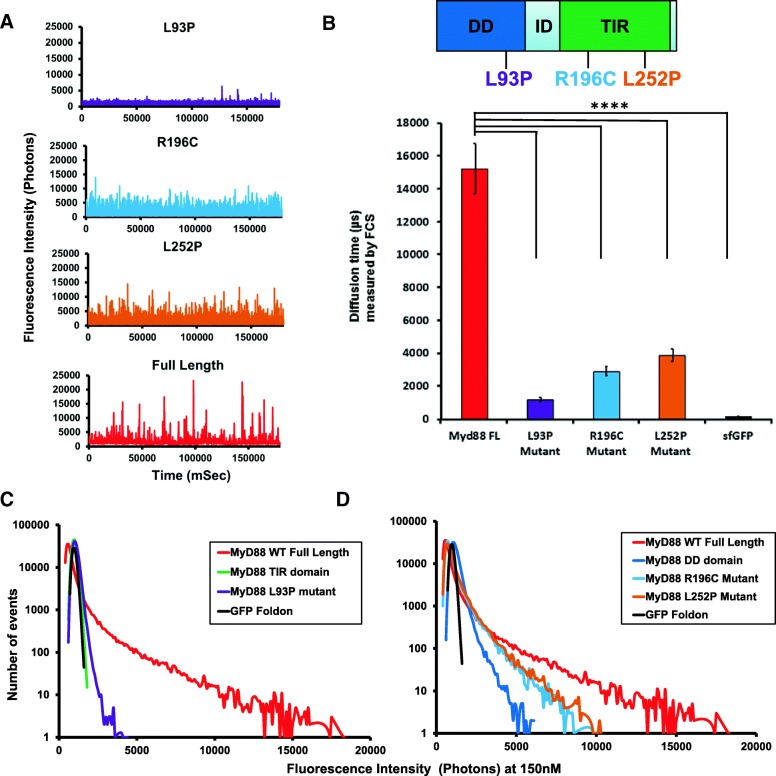
Fig. 4.
Disease-associated point mutations abrogate domain function and thus, MyD88 polymerisation. a Fluorescence time-traces obtained with disease-associated point mutants of the full-length MyD88 protein, as well as the wild-type full-length MyD88 at a protein concentration of 150 nM. As in Fig. 1, the diffusion of an oligomer equates to the same number of fluorophores moving through the confocal volume, creating a burst of fluorescence in the time-trace being directly proportional to the size of the oligomer. b Diffusion time (μs) measured by FCS showing the drastic shift in diffusion time when comparing the mutants to the wild-type protein. c Fluorescence intensity histogram showing that the L93P point mutation, which is within the DD, in GFP-tagged MyD88 renders the polymerisation propensity similar to the MyD88 TIR domain alone. d Fluorescence intensity histogram demonstrating that the R196C and L252P point mutations (present within the TIR domain) in the GFP-tagged MyD88 render the polymerisation propensity more similar to the MyD88 DD alone. Fluorescence time-traces and intensity histograms in a, c and d are representative of eight independent experiments. Values in b are ±SD from these eight measurements. Sidak’s multiple comparisons test (****P < 0.0001)