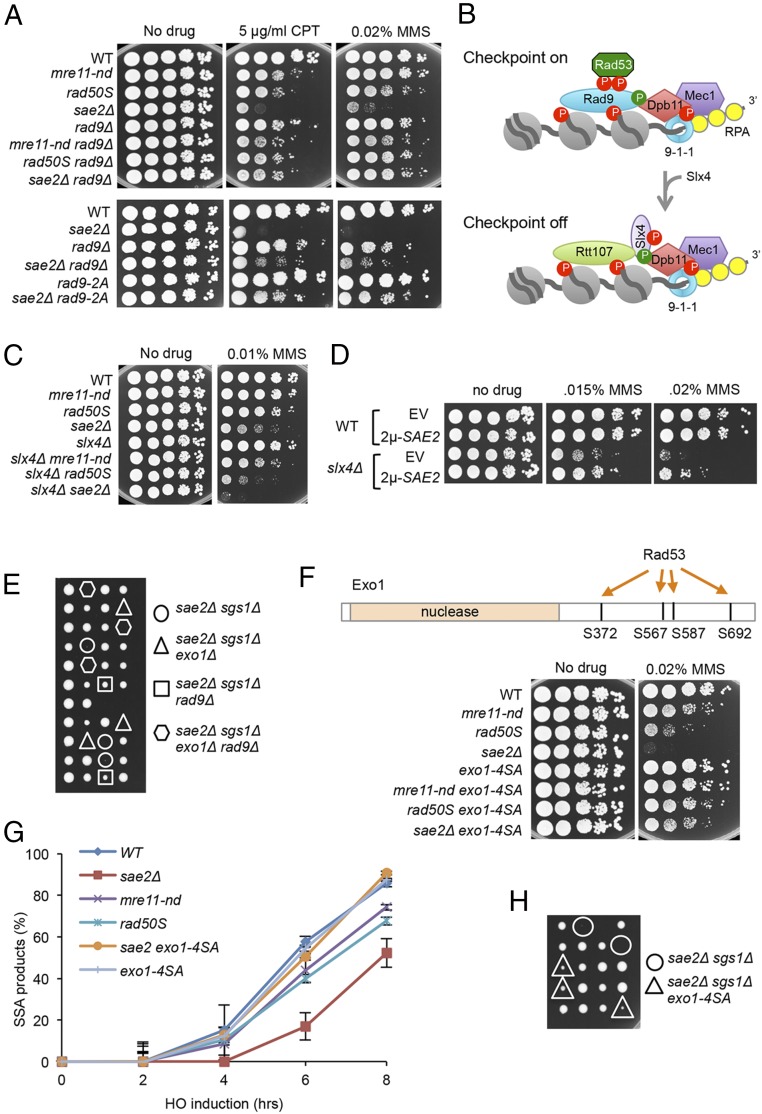
Fig. 3.
Rad9 chromatin binding and Rad53 activation contribute to sae2Δ DNA damage sensitivity and sae2Δ sgs1Δ lethality. (A) Ten-fold serial dilutions of the indicated strains spotted on plates without drug, or plates containing indicated DNA damaging agents. (B) Schematic showing stabilization of Rad9 binding to chromatin by Dpb11 to activate the DNA damage checkpoint. CDK (green circles)- and Mec1 (red circles)-phosphorylated Slx4 competes with Rad9 for Dpb11 interaction, dampening the checkpoint. (C) Ten-fold serial dilutions of the indicated strains spotted on plates without drug, or plates containing indicated DNA damaging agents. (D) Ten-fold serial dilutions of the indicated strains spotted on plates without drug, or plates containing MMS. (E) Diploids heterozygous for the indicated mutations were sporulated and tetrads dissected on YPD plates. (F) Upper schematic showing Rad53-dependent phosphorylation sites in the C-terminal region of Exo1. Lower shows 10-fold serial dilutions of the indicated strains spotted on plates without drug, or plates containing indicated DNA damaging agents. (G) SSA kinetics were assessed by qPCR of genomic DNA from the indicated strains before and after HO induction. Error bars indicated SD from three independent trials. (H) Diploids heterozygous for the indicated mutations were sporulated and tetrads dissected on YPD plates.