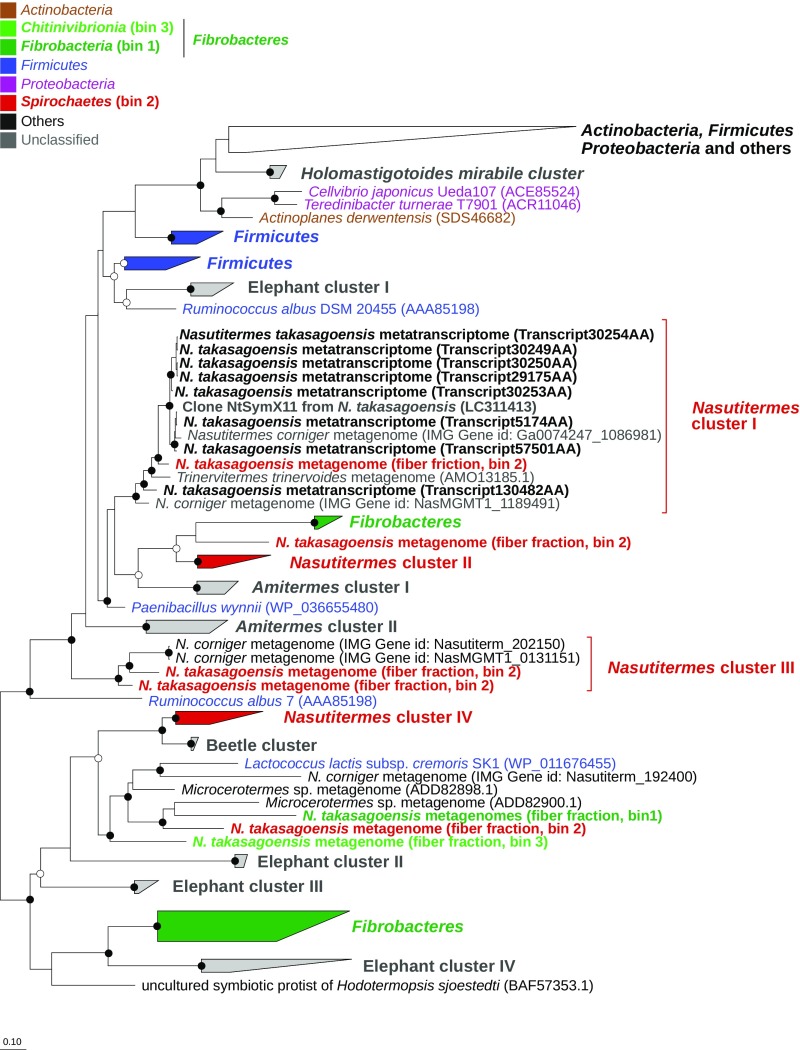
Fig. 5.
Phylogenetic relationship among bacterial GH11 members and phylum-level classification of homologs from fiber-associated bacteria in the hindgut of N. takasagoensis. The unrooted tree is based on 164 alignment positions and represents a consensus phylogeny obtained by using maximum-likelihood (ML) and Bayesian (BA) inference and the WAG model of protein evolution (60). Sequences obtained in this study are shown in bold, and text color indicates the taxonomic affiliation of homologs obtained from bacterial genomes and compositional bins. Node support was determined with the χ2 approximate likelihood ratio test (i.e., ML) and posterior probability (i.e., BA); confidence values are indicated by circles (open, ≥90% support in at least one method; filled, ≥90% support in both methods). The detailed tree, including all accession numbers and confidence values, is provided in SI Appendix, Fig. S5.