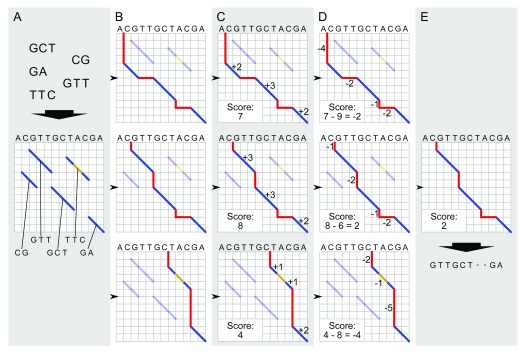
Figure 2. Global pairwise alignment schematic.
Schematic of modified Needleman-Wunsch algorithm for global alignment of an input contig to a reference genome. The process starts with local alignments between the contig and the reference genome (blue diagonals in A). All possible combinations of these local alignments are enumerated by realizing all paths connecting contigs from the upper left to lower right corner of the matrix ( B). Each alignment is scored: matches contribute positive scores (dark blue lines in C), while indels (red) and mismatches (gold) incur a penalty ( D). The alignment with the highest score is selected as the best global alignment ( E) for the next step in graph genome creation; ties among global alignments are resolved arbitrarily.