FIGURE 1.
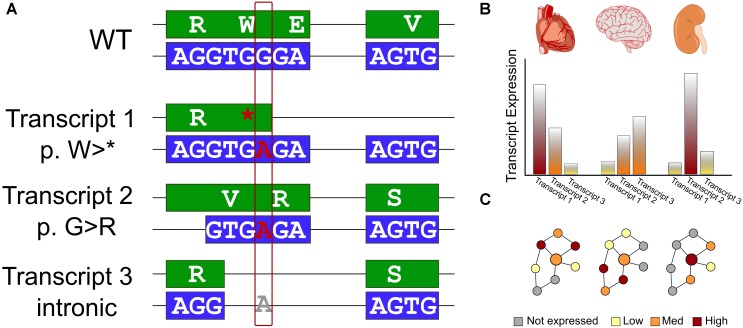
Interpreting the effects of genomics variants identified in WES requires transcript selection and context. (A) Schematic of a single genomic variant within a protein-coding gene that has a different impact in each of three isoforms. We first show a reference WT sequence with exons shown in boxes and introns as connecting lines. The DNA sequence is colored blue and encoded protein sequence in green. We consider the effect on this example sequence of a G > A variant. In the first transcript, the variant introduces an early stop codon. The second, an alternative splice site is used leading to a different reading frame, and a missense variant. Finally, for a third transcript an additional splicing pattern skips over the altered region and the genomic variant has no effect on the encoded protein sequence. (B) Many genes exhibit variability in expression level across tissues. This must also be considered for an accurate assessment of the effect of genomic variants. (C) Data interpretation is further challenged by the reality of biologic networks – the effects of transcript selection and tissue-specific expression affect not only the gene of interest (represented by a larger circle), but also the genes interacting with it (represented by smaller circles) in biologic networks or protein complexes. Thus, even if these features do not directly affect a gene of interest, a genomic variant may have a different apparent functional effect due to different environments in different biologic contexts.