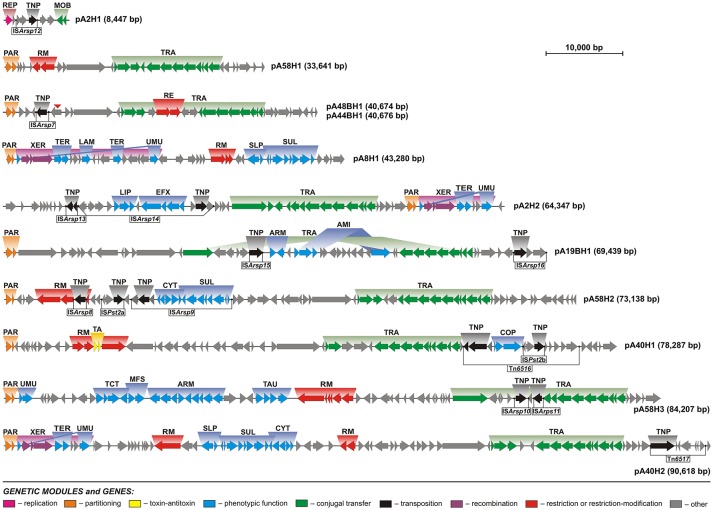
Figure 2.
Linear maps showing the genetic structure and organization of the circular plasmids of Antarctic Arthrobacter spp. Arrows indicate genes and their transcriptional orientation. Predicted genetic modules are indicated by colored boxes: ARM, aromatic compound utilization; COP, copper resistance; CYT, cytochrome biogenesis; EFX, drug-specific efflux; LAM, laminarinase; LIP, lipoprotein export system; MFS, major facilitator transporter; MOB, mobilization for conjugal transfer; PAR, partitioning; RE, type IV restriction enzyme; REP, replication; RM, restriction-modification; TA, toxin-antitoxin; TNP, transposition; SLP, sulfate transport; SUL, sulfur metabolism; TAU, taurine transport; TCT, tricarboxylate transport; TER, tellurium resistance; TRA, conjugal transfer; UMU, UV-damage protection/repair; XER, recombination. Potentially active transposable elements were distinguished and the location of their IR sequences is marked by black dots. A red triangle indicates the gene that was disrupted in the plasmid pA44BH1, but is intact within pA48BH1.