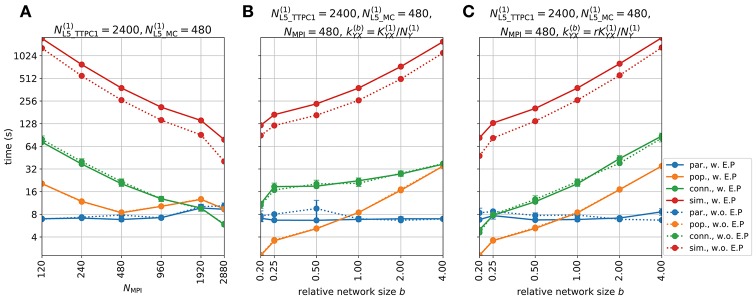
Figure 7.
Parallel performance with networks in LFPy. (A) Initialization of parameters (par.), population create (pop.), connectivity build (conn.) and main simulation time (sim.) as functions of number of physical CPU cores/MPI processes (NMPI). The reference network population sizes for X ∈ {L5_TTPC1, L5_MC} are given in the panel title. The network was otherwise constructed with synapse, stimulus and connectivity parameters for each possible connection as given in Tables 1–3. Times shown with continuous lines were obtained for simulations that included calculations of extracellular potentials and current dipole moments as in Figures 2–6 (w. E.P.), while times shown with dotted lines were obtained for simulations with no such signal predictions (w.o. E.P.). Each data value is shown as the mean and standard deviation of times obtained from N = 3 network realizations instantiated with different random seeds. (B) Initialization of parameters, population create, connectivity build and main simulation time as functions of network size relative to the reference network population sizes for X ∈ {L5_TTPC1, L5_MC} as given in the panel title. The superset “(1)” denotes a relative network size b = 1. Simulations were run using a fixed MPI process count NMPI and connection probabilities were recomputed for different values of b, such that the expected total number of connections was constant between each simulation (using 20). The set-up was otherwise identical to the set-up in (A). (C) Same as (B), but with a fixed expected per-cell synapse in-degree across different relative network sizes.