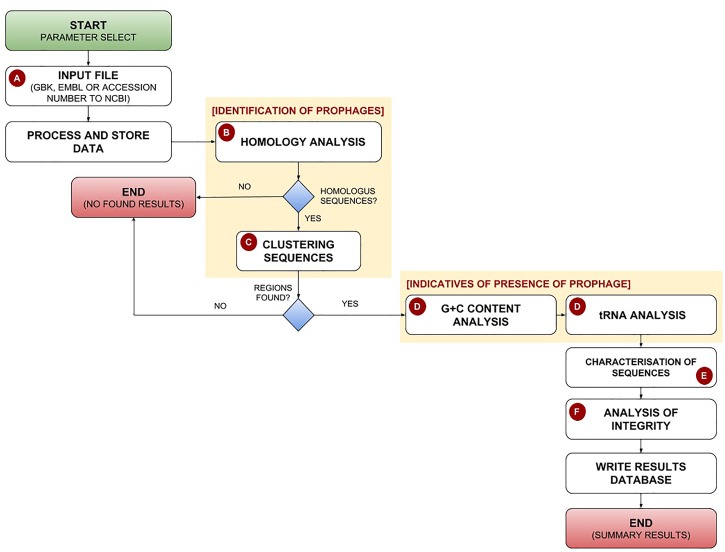
FIGURE 1.
Pipeline for identification and characterization of prophages by the PhageWeb computational tool. (A) The pipeline receives as input the parameters (Alignment Identity and MinPts – minimum number of phage proteins in a region) and the annotation file (GenBank, Embl or the NCBI’s Accession Number) of the target genome which will be evaluated; (B) The homology search of the coding sequences in a local database based on the publically available sequences annotated as phage obtained from NCBI, which is automatically updated once per week; (C) After identifying the homology sequences, a clustering analysis based on the distance of the elements is performed: part of phage region to be evaluated on the amount of prophage there; (D) This optional step is useful to know if the identified region has more features which can be an evidence of prophage: G+C content (due to be possible variation of G+C content on the flanks of the prophage) and the presence of tRNAs on the flank; (E) Use of web services to connect biological databases to perform the functional characterization of the identified sequences; (F) Verification of the probable integrity of the prophage based on the composition of genes of each identified phage.