Figure 1.
Arabidopsis LUC7 Proteins Redundantly Control Plant Development.
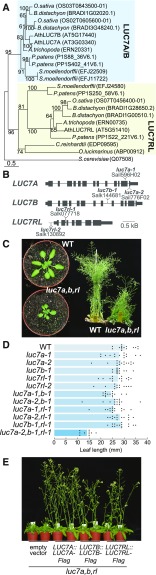
(A) Phylogenetic analysis of LUC7 proteins in the plant kingdom using Saccharomyces cerevisiae as an external group. Sequences were aligned by Muscle and Maximum likelihood (PhYML) with 1000 bootstraps.
(B) Exon/intron structure of Arabidopsis LUC7A, LUC7B, and LUC7RL. Dotted lines indicate the positions of T-DNA insertions.
(C) Wild-type and luc7a-2 luc7b-1 luc7rl-1 (luc7a-2,b-1,rl-1) mutant plants growing for 21 d in long-day conditions (left) and for 45 d in standard greenhouse conditions (right).
(D) Length of the longest rosette leaf of 21-d-old wild-type, luc7 single, double, and triple mutant plants growing under long-day conditions. Leaves of 10 to 15 individual plants were measured. Dots indicate individual data points.
(E) Complementation of luc7a-2 luc7b-1 luc7rl-1 mutant by LUC7A, LUC7B, and LUC7RL genomic rescue constructs. Transformation of an “empty” binary vector served as a control. Two independent transgenic lines for each construct are shown.