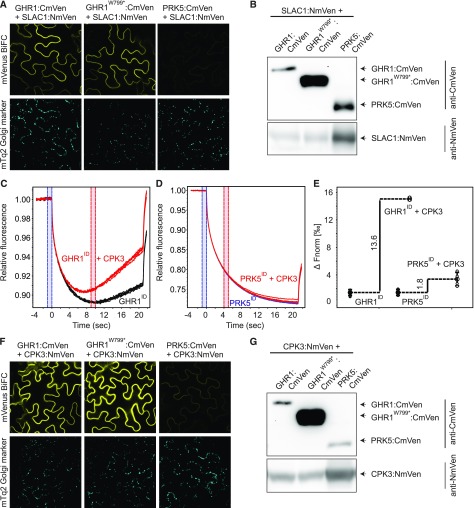
Figure 8.
Interaction of GHR1 with SLAC1 and CPK3.
(A) and (F) Bimolecular fluorescence complementation assays were performed with N. benthamiana leaves infiltrated with 35S:GHR1:CmVen, 35S:GHR1W799*:CmVen or 35S:PRK5:CmVen with 35S:SLAC1:NmVen or 35S:CPK3:NmVen. mVenus (top row) and mTq2 (bottom row) signals are shown. Expression of the mTq2 Golgi marker (bottom row) confirms that transformation of all constructs was successful. N. benthamiana leaf tissue was imaged 48 h after transformation. Identical settings were used for all constructs allowing direct comparison. All experiments were repeated at least three times, and representative BiFC images are shown.
(B) and (G) Immunoblots showing expression of split-YFP fusion proteins in BiFC samples used for confocal imaging.
(C), (D), and (E) Analysis of in vitro binding of GHR1ID and PRK5ID with CPK3. Interaction between intracellular domain of GHR1 (GHR1ID) and PRK5 (PRK5ID) fused to GST with CPK3 was monitored by microscale thermophoresis (MST). (C) MST traces of 66 nM fluorescently labeled GHR1ID in the absence (black) and presence (red) of 5 µM CPK3. (D) MST traces of 40 nM fluorescently labeled PRK5ID in the absence (blue) and presence (red) of 5 µM CPK3. (E) Difference in normalized fluorescence between GHR1ID or PRK5ID alone and in the presence of CPK3 as indicated. All experiments were performed four times.