FIGURE 2.
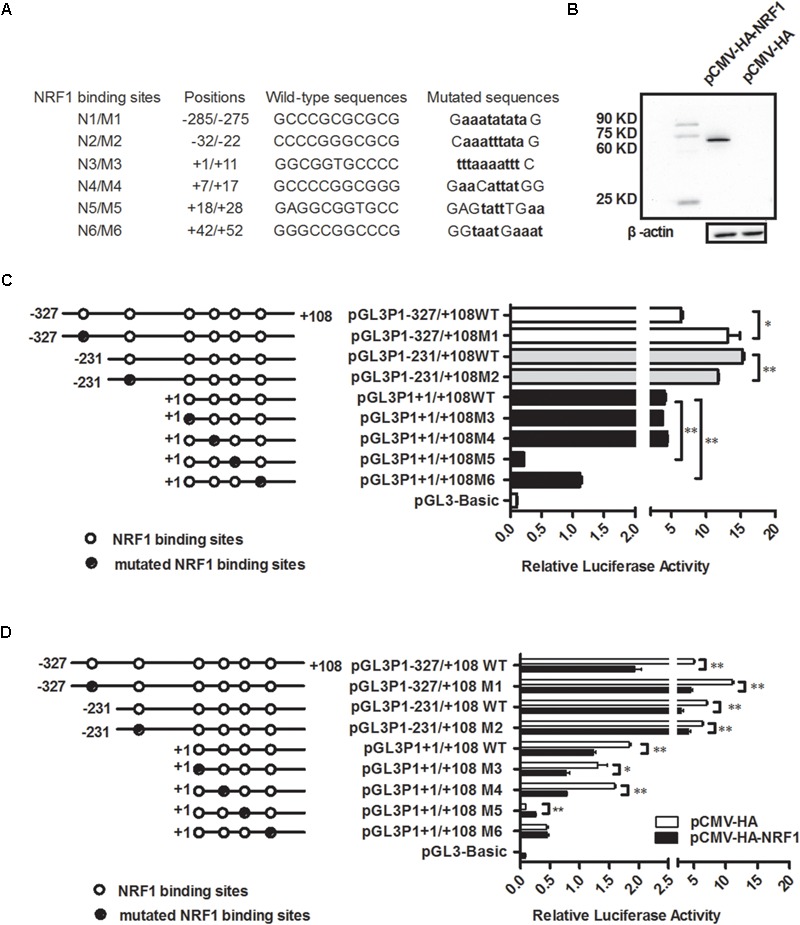
NRF1 represses P1 promoter activity. (A) The nucleotide sequences of the six putative NRF1 binding sites and their respective mutations in the P1 promoter. The P1 promoter mutants were created by direct DNA synthesis and subsequent cloning, and the positions of the six putative NRF1 binding sites in the P1 promoter region were numbered relative to the transcription start site (TSS). NRF1 binding sites are indicated by capital letters, and their mutated nucleotides are indicated by bold lowercase letters. (B) Western blot identification of chicken NRF1 expression vector (pCMV-HA-NRF1). The pCMV-HA-NRF1 or empty pCMV-HA vector was transfected into DF1 cells. The cell lysates were harvested 48 h after transfection and immunoblotted with an HA-specific antibody. (C) The effects of mutation of the six individual putative NRF1 binding sites on basal P1 promoter activity. The indicated P1 promoter constructs (pGL3P1-327/+108, pGL3P1-231/+108, and pGL3P1+1/+108) or indicated P1 promoter mutant constructs (pGL3P1-327/+108M1, pGL3P1-231/+108M2, pGL3P1+1/+108M3, pGL3P1+1/+108M4, pGL3P1+1/+108M5, and pGL3P1+1/+108M6), along with pRL-TK, were cotransfected into DF1 cells, and luciferase activity was determined 48 h after transfection. The open circles indicate the wild-type NRF1 binding sites, and the filled circles indicate the mutated NRF1 binding sites. All data represent mean ± SE. Statistical significance was determined by Student’s t-test comparing mutated versus wild-type NRF1 binding site. ∗p < 0.05, ∗∗p < 0.01. (D) Effects of mutation of the six individual NRF1 binding sites on NRF1-mediated inhibition of the P1 promoter. DF1 cells were cotransfected with the indicated reporter constructs along with pCMV-HA-NRF1 or empty pCMV-HA vector and pRL-TK. Luciferase activity was determined 48 h after cotransfection. All data represent the mean ± SE. Statistical significance was determined by Student’s t-test comparing the cotransfection of the designated reporter constructs and pCMV-HA-NRF1 versus the cotransfection of the designated reporter constructs and empty pCMV-HA vector. ∗p < 0.05, ∗∗p < 0.01.
