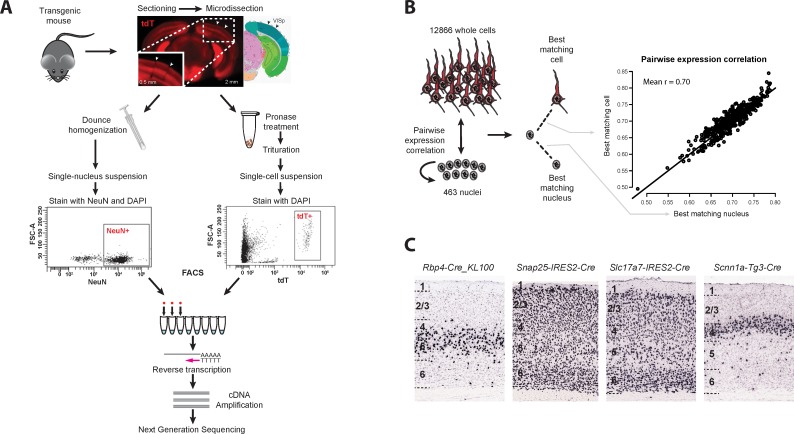
Fig 1. Identification of an expression-matched set of single nuclei and whole cells from mouse primary visual cortex (VISp).
(A) Whole brains were dissected from transgenic mice, sectioned into coronal slices, and individual layers of VISp were microdissected. Nuclei were dissociated from layer 5, stained with DAPI and against the neuronal marker NeuN. Single NeuN-positive nuclei were isolated by fluorescence-activated cell sorting (FACS). In parallel, whole cells were dissociated from all layers, and single td-Tomato-positive cells were isolated from multiple different Cre-driver lines. Single nucleus and cell mRNA were reverse-transcribed, amplified, and sequenced to measure genome-wide gene expression levels. (B) Left: 463 nuclei from layer 5 and 12,866 whole cells from all layers, which passed quality control metrics were used to determine expression correlation between each nucleus and every other nucleus and cell. Expression similarity can vary based on sample quality, so nuclei were compared to each other to provide a baseline expected similarity. For each nucleus, the best matching nucleus and cell were selected based on maximal correlation. Right: Cells and nuclei displayed comparable expression similarities to all nuclei, with average correlation equal to 0.70 and 95% of correlations between 0.63 and 0.78. This suggested that nuclei and cells were well matched. (C) Chromogenic RNA in situ hybridization (ISH) for tdTomato mRNA in VISp of transgenic mice (Cre-lines crossed to Ai14 Cre reporter [21]). Shown are the tissue sections from 4 Cre-driver lines from which the majority of the best-matching cells to L5 nuclei were derived. As expected, all Cre-lines label cells in layer 5 and adjacent layers.